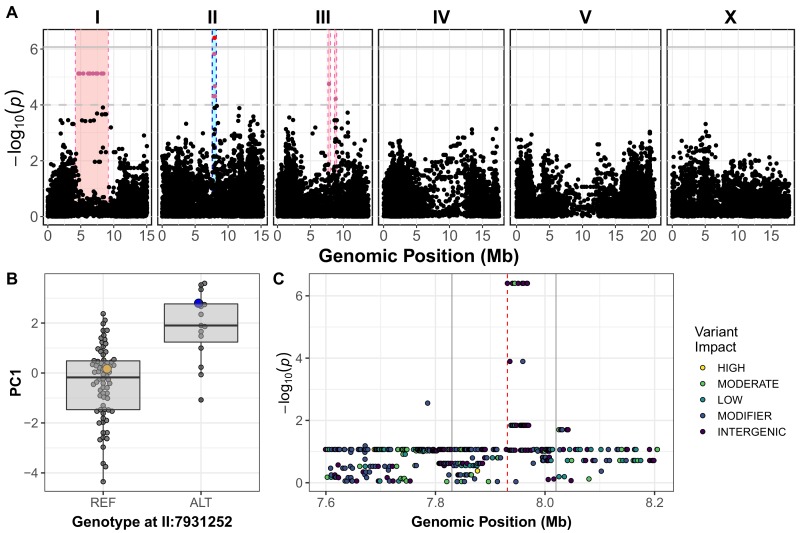
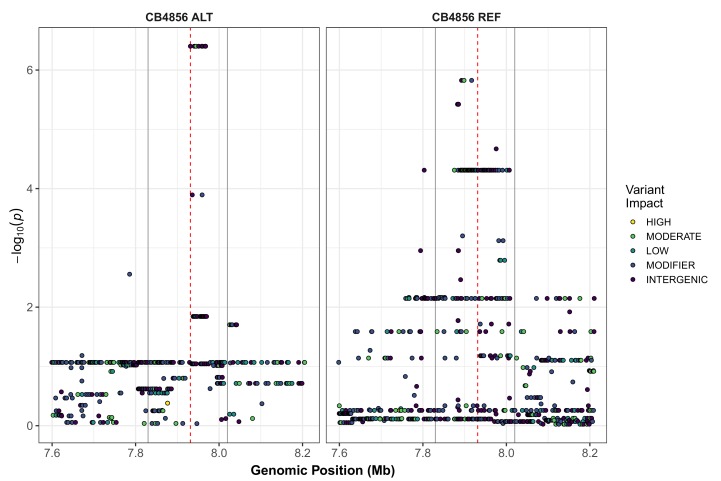
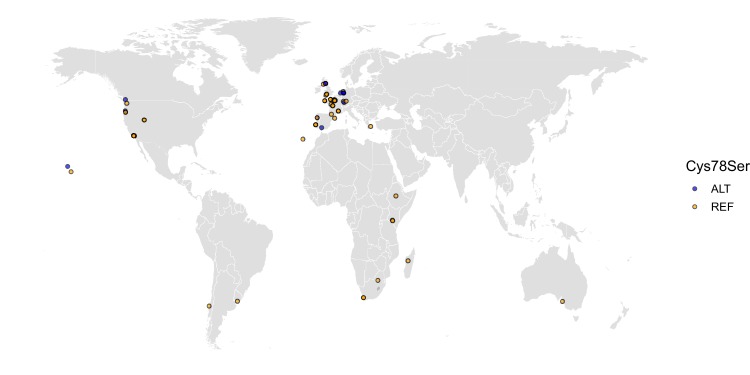
Figure 2. Variation in C. elegans wild isolates responses to arsenic trioxide maps to the center of chromosome II.
(A) A manhattan plot for the first principal component in the presence of 1000 µM arsenic trioxide is shown. Each dot represents an SNV that is present in at least 5% of the assayed wild population. The genomic position in Mb, separated by chromosome, is plotted on the x-axis and the -log10(p) for each SNV is plotted on the y-axis. SNVs are colored red if they pass the genome-wide Bonferroni-corrected significance (BF) threshold, which is denoted by the gray horizontal line. SNVs are colored pink if they pass the genome-wide eigen-decomposition significance (ED) threshold, which is denoted by the dotted gray horizontal line. The genomic region of interests surrounding the QTL that pass the BF and ED thresholds are represented by cyan and pink rectangles, respectively. (B) Tukey box plots of phenotypes used for association mapping in (A) are shown. Each dot corresponds to the phenotype of an individual strain, which is plotted on the y-axis. Strains are grouped by their genotype at the peak QTL position (red SNV from panel A, ChrII:7,931,252), where REF corresponds to the allele from the reference N2 strain. The N2 (orange) and CB4856 (blue) strains are highlighted. (C) Fine mapping of the chromosome II region of interest (cyan region from panel A, 7.60–8.21 Mb) is shown. Each dot represents an SNV present in the CB4856 strain. The association between the SNV and first principal component is shown on the y-axis and the genomic position of the SNV is shown on the x-axis. Dots are colored by their SnpEff predicted effect.