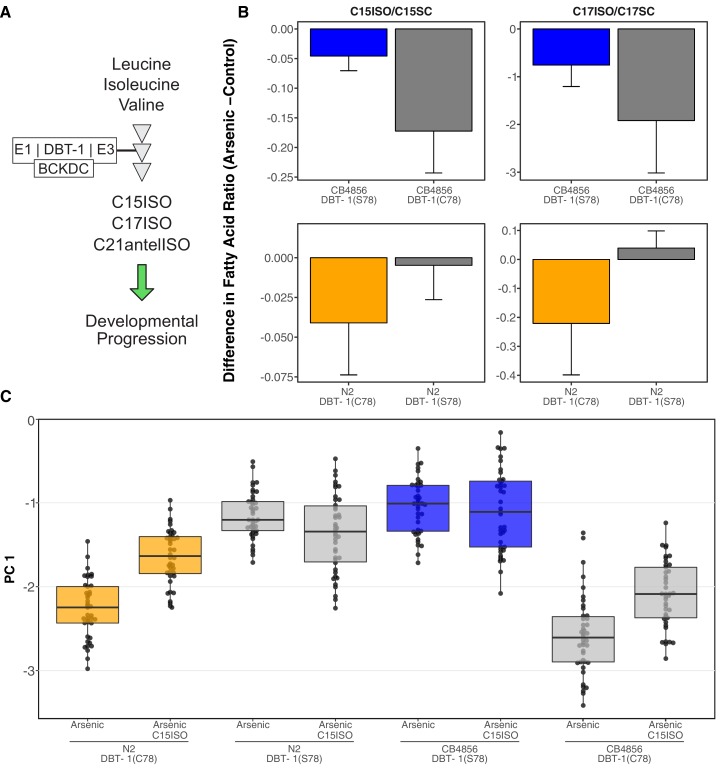
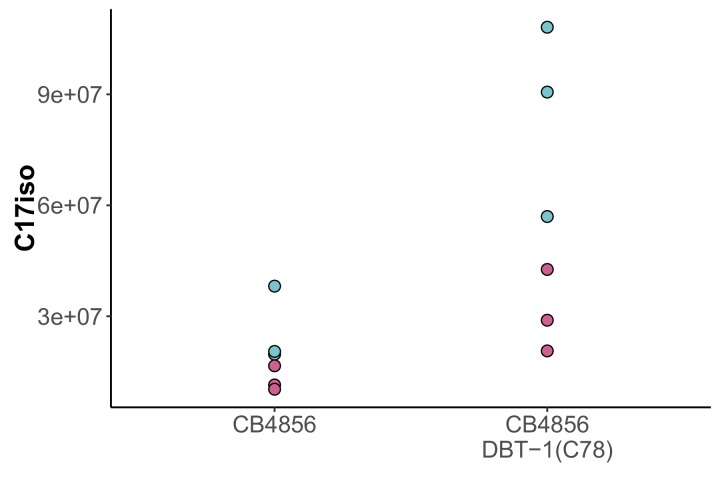
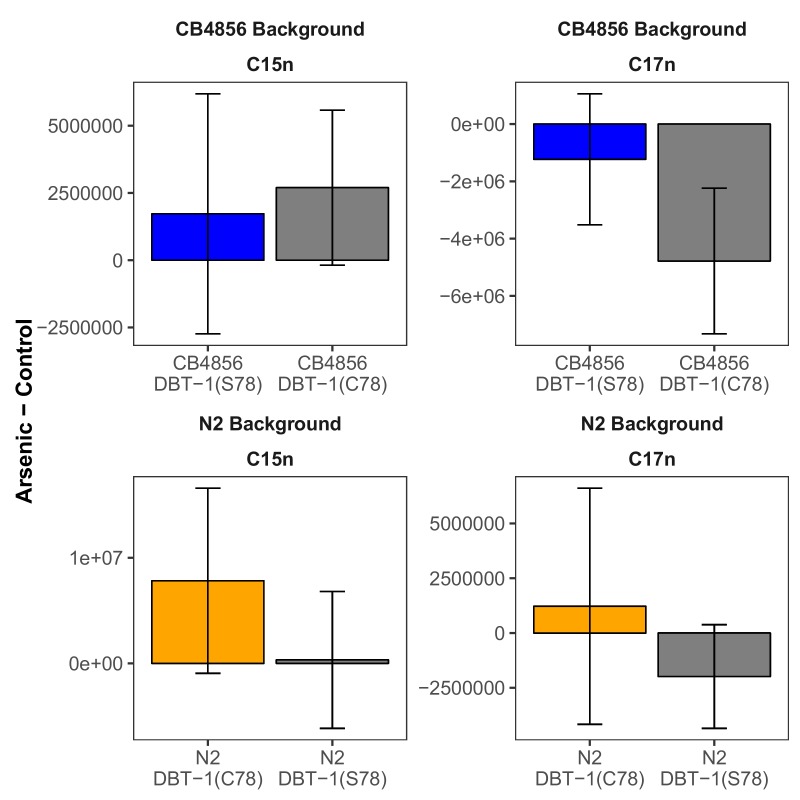
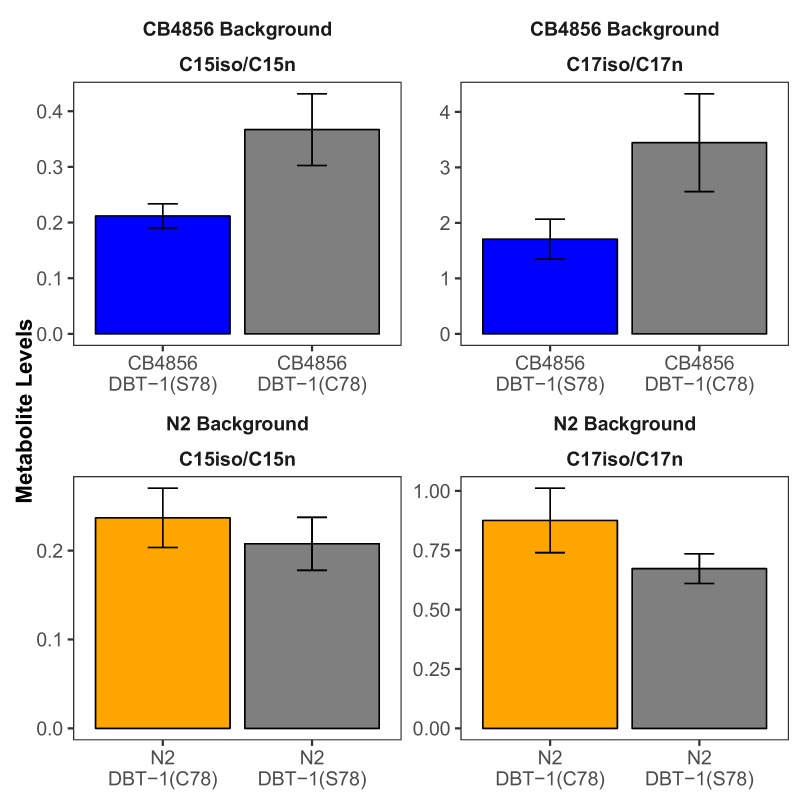
Figure 4. Differential production of mmBCFA underlies DBT-1(C78)-mediated sensitivity to arsenic trioxide.
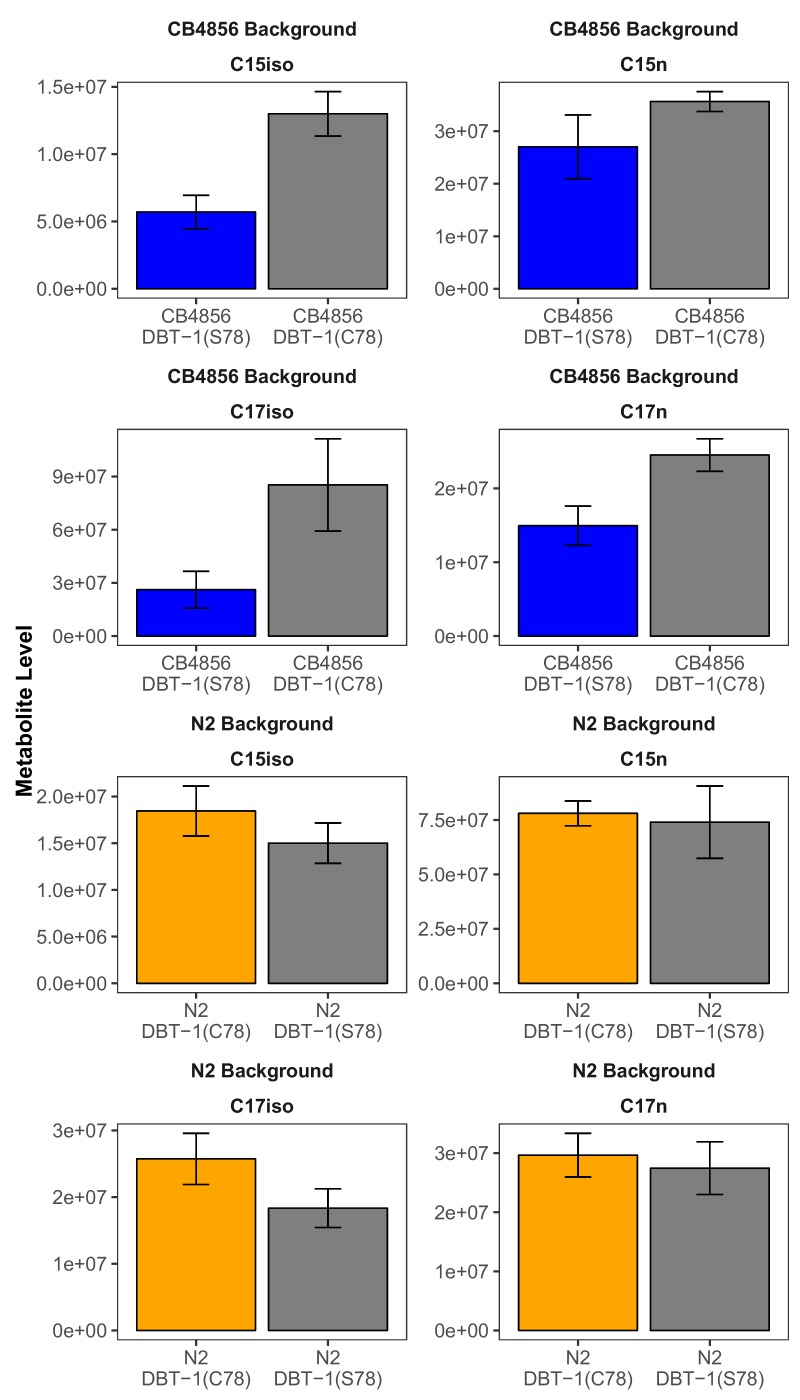
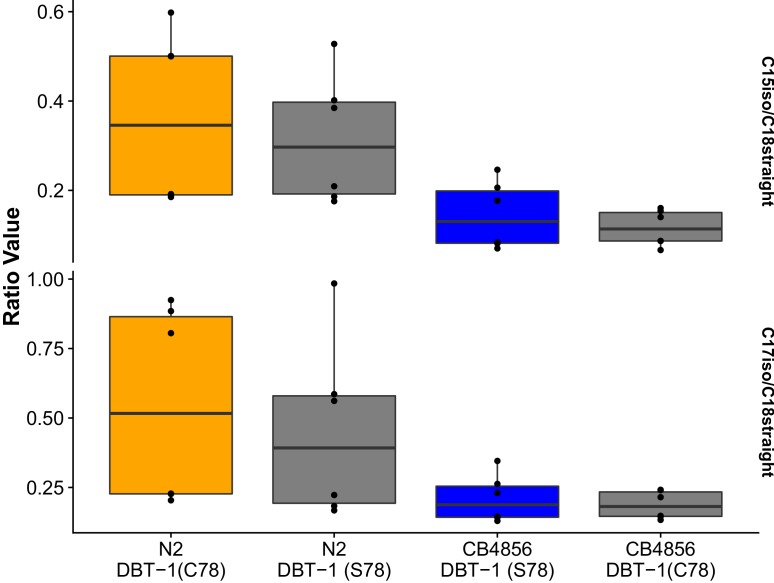
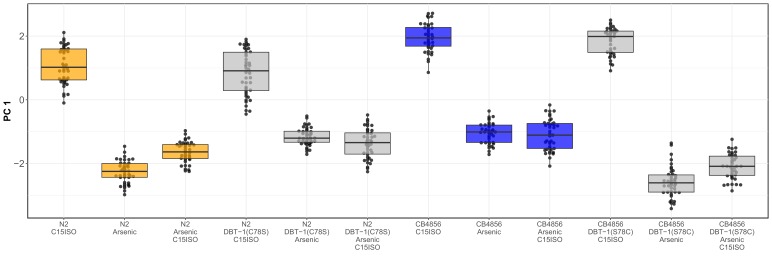
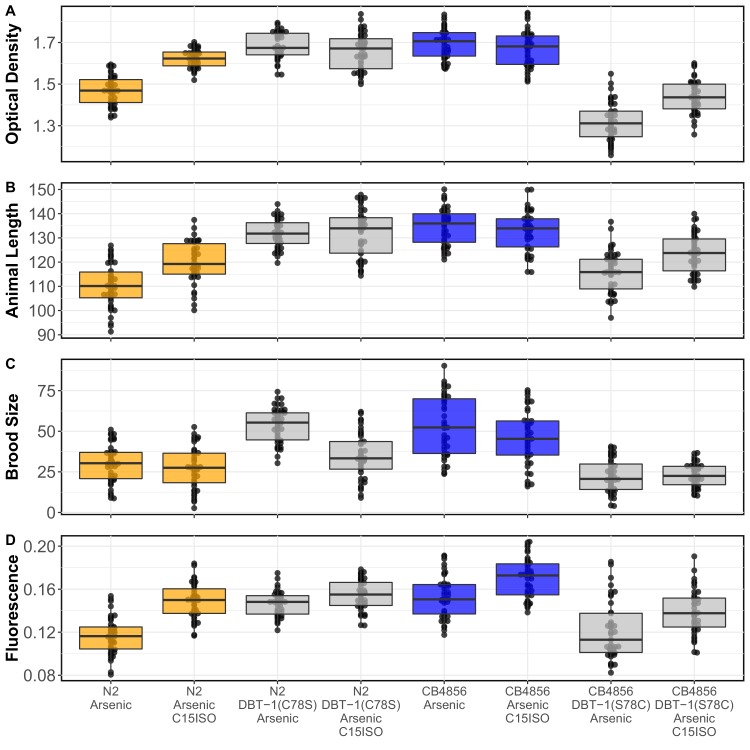
(A) A simplified model of BCAA catabolism in C. elegans. The BCKDH complex, which consists of DBT-1, catalyzes the irreversible oxidative decarboxylation of branched-chain ketoacids. The products of thesebreakdown can then serve as building blocks for the mmBCFA that are required for developmental progression. (B) The difference in the C15ISO/C15SC (left panel) or C17ISO/C17SC (right panel) ratios between 100 μM arsenic trioxide and control conditions is plotted on the y-axis for three independent replicates of the CB4856 and CB4856 allele replacement strains and six independent replicates of the N2and N2 allele replacement strains. The difference between the C15 ratio for the CB4856-CB4856 allele replacement comparison is significant (Tukey HSD p-value = 0.0427733), but the difference between the C17 ratios for these two strains is not (Tukey HSD p-value = 0.164721). The difference between the C15and C17 ratios for the N2-N2 allele replacement comparisons are both significant (C15: Tukey HSD p-value = 0.0358; C17: Tukey HSD p-value = 0.003747). (C) Tukey box plots median animal length after arsenic trioxide or arsenic trioxide and 0.64 μM C15ISO exposure are shown (N2, orange; CB4856, blue; allele replacement strains, gray). Labels correspond to the genetic background and the corresponding residue at position 78 of DBT-1 (C for cysteine, S for serine). Every pair-wise strain comparison is significant except for the N2 DBT-1(S78) - CB4856 comparisons (Tukey’s HSD p-value < 1.43E-6).