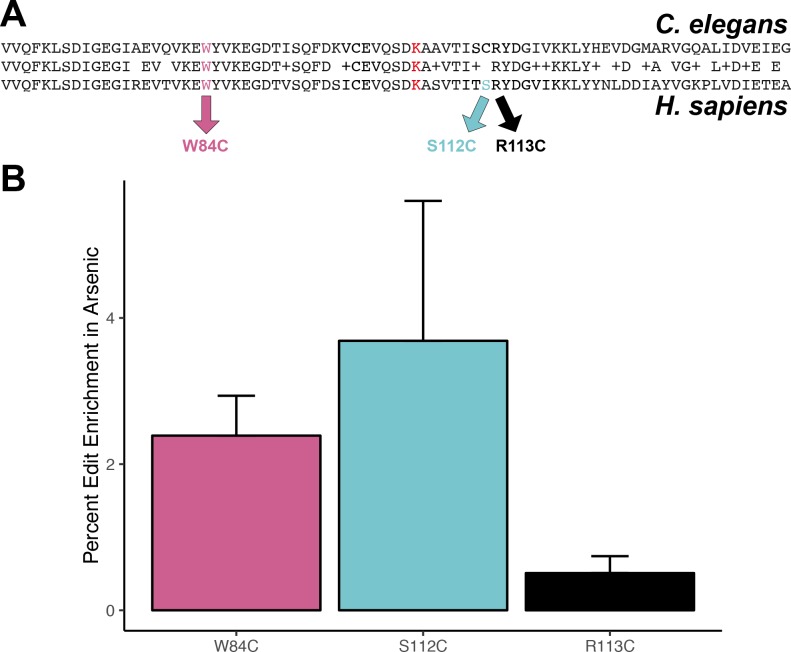
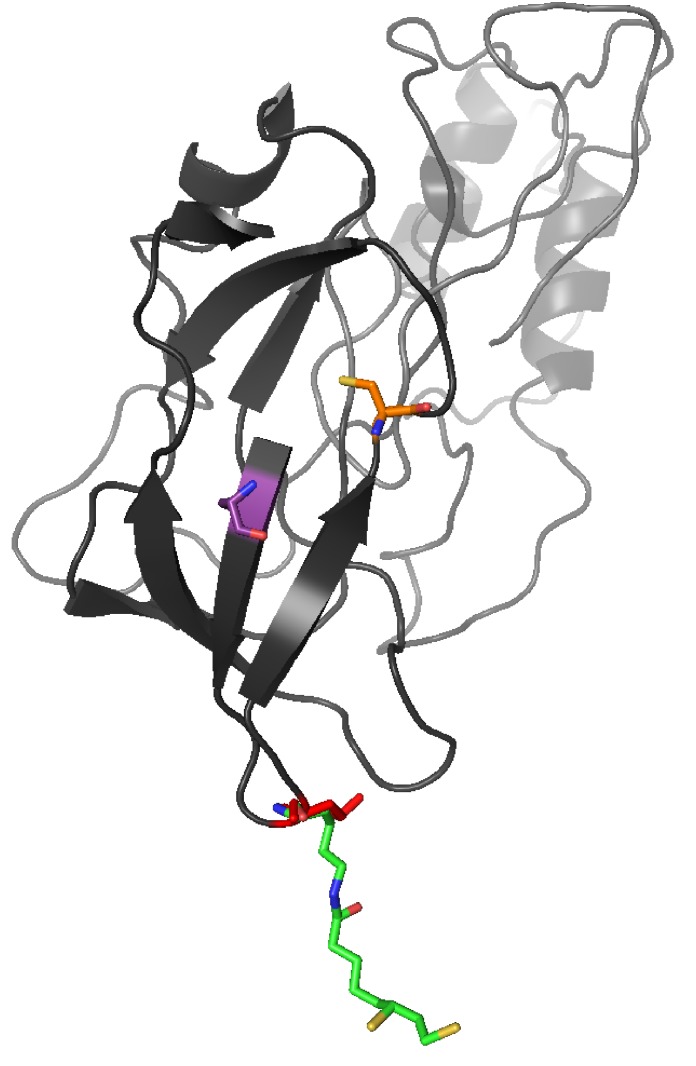
Figure 5. Protective effect of cysteine residues in human DBT1.
(A) Alignment of C. elegans DBT-1 and H. sapiens DBT1. The residues tested for an arsenic-specific effect are indicated with arrows - W84C (pink), S112C (blue), and R113C (black). The lysine that is post-translationally modified with a lipoid acid is highlighted in red. (B) The percent increase of edited human cells that contain the W84C, S112C, or R113C amino acid change in DBT1 in the presence 5 µM arsenic trioxide relative to control conditions are shown. The number of reads in 5 µM arsenic trioxide for all replicates are significantly different from control conditions (Fisher’s exact test, p-value<0.011).