Abstract
Hepatitis B virus (HBV) replicates its DNA genome through reverse transcription of an RNA intermediate. The lack of proofreading capacity of the viral DNA polymerase results in a high mutation rate of HBV genome. Under the selective pressure created by the nucleos(t)ide analogue (NA) antiviral drugs, viruses with resistance mutations are selected. However, the replication fitness of NA-resistant mutants is markedly reduced compared to wild-type. Compensatory mutations in HBV polymerase, which restore the viral replication capacity, have been reported to arise under continuous treatment with lamivudine (LMV). We have previously identified a highly replicative LMV-resistant HBV isolate from a chronic hepatitis B patient experiencing acute disease exacerbation. Besides the common YMDD drug-resistant mutations, this isolate possesses multiple additional mutations in polymerase and core regions. The transcomplementation assay demonstrated that the enhanced viral replication is due to the mutations of core protein. Further mutagenesis study revealed that the P5T mutation of core protein plays an important role in the enhanced viral replication through increasing the levels of capsid formation and pregenomic RNA encapsidation. However, the LMV-resistant virus harboring compensatory core mutations remains sensitive to capsid assembly modulators (CpAMs). Taken together, our study suggests that the enhanced HBV nucleocapsid formation resulting from core mutations represents an important viral strategy to surmount the antiviral drug pressure and contribute to viral pathogenesis, and CpAMs hold promise for developing the combinational antiviral therapy for hepatitis B.
Keywords: Hepatitis B virus, drug resistance, replication fitness, capsid assembly
Introduction
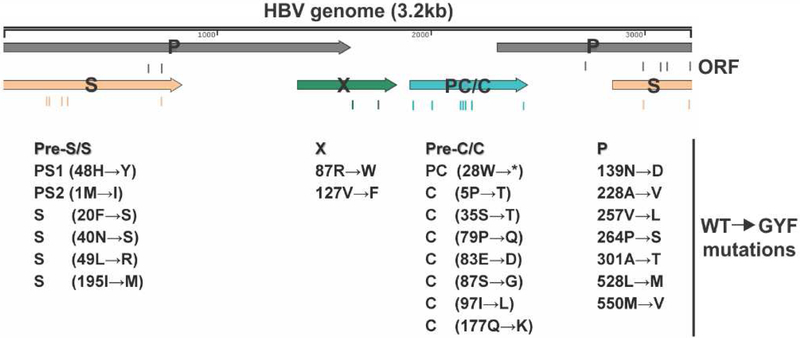
Hepatitis B virus (HBV) is a medically important human pathogen which causes chronic infection in approximately 257 million people worldwide, leading to a series of life-threatening complications, including cirrhosis, liver failure and hepatocellular carcinoma (Alter et al., 2018). HBV is a partially double stranded DNA virus that replicates its genome through reverse transcription of a pregenomic (pg) RNA (Block et al., 2007). HBV replication can be suppressed effectively by nucleos(t)ide analogue (NA) reverse transcriptase (RT) inhibitors, which target the viral polymerase (pol) and cease DNA chain elongation. Unfortunately, the emergence of resistant viral mutants significantly limits the effectiveness of some NAs (Gish et al., 2012; Lampertico and Liaw, 2012). Usually, the NA-resistance mutations in the catalytic YMDD motif of pol lead to a decreased viral replication due to reduced enzymatic activity of the mutant pol (Allen et al., 1998; Sheldon et al., 2006). However, compensatory mutations have been reported to partially restore the viral replication capacity, including rtL80V/I, rtL82M, rtV173L and rtV207I, mostly in the RT region of pol (Delaney et al., 2003; Ono et al., 2001; Pichoud et al., 1999). In addition, a NA-resistant mutant HBV with increased replication has also been found in chronic hepatitis B (CHB) patients with progressive liver disease (Zoulim and Locarnini, 2009). We have previously identified a highly replicative lamivudine (LMV)-resistant HBV isolate from CHB patients experiencing fulminant hepatitis, which, in addition to the YMDD mutation, possesses multiple mutations in pol, core, X, and surface genes (Zhang et al., 2005) (Fig. 1).
Figure 1. The lamivudine-resistant GYF isolate harbors multiple mutations.
The one unit-length HBV genome is schematically illustrated in linear form and the encoded ORFs are shown with arrows, including the surface (S) proteins ORF, polymerase (P) ORF, precore/core (PC/C) ORF, and HBx (X) ORF. The mutations carried by GYF isolate are listed underneath their corresponding ORFs as amino acid changes from the wildtype (WT) sequence at the indicated positions.
HBV core protein or hepatitis B core antigen (HBcAg) plays a critical role in HBV life cycle, which self-assembles into the capsid of virus and encapsdiates viral pgRNA and pol to form the cytoplasmic nucleocapsid (Bartenschlager and Schaller, 1992; Birnbaum and Nassal, 1990), inside of which pol synthesizes HBV DNA by reverse transcribing pgRNA (Summers and Mason, 1982). HBV core protein is composed of 183 amino acids, of which the first N-terminal 149 residues are characterized as the assembly domain that mediates the capsid formation (Hu and Liu, 2017). HBcAg possesses a four-helix bundle structure, two core monomers bind together to form a dimer first, followed by the construction of viral capsid from 90 or 120 copies of dimer (Birnbaum and Nassal, 1990; Bottcher et al., 1997; Wynne et al., 1999). Naturally-occurring mutations within the N-terminus of HBcAg have been reported to affect capsid assembly, uncoating, or virion secretion (Cui et al., 2015; Ning et al., 2018; Pairan and Bruss, 2009). Among those core mutations with high frequencies in CHB patients, the switch of phenylalanine (F) or isoleucine (I) to leucine (L) at residue 97 results in an enhanced immature secretion phenotype, in which the virion mainly contains single-stranded (SS) HBV DNA rather than the mature relaxed circular (rc) DNA (Ceres et al., 2004; Suk et al., 2002; Yuan et al., 1999a; Yuan et al., 1999b). Codon 5 is another hot spot of naturally occurring mutation of HBcAg. Previous studies have shown that the substitution of proline (P) with threonine (T) at codon 5 of HBcAg caused lower levels of virion secretion, which, would revert to the wild type secretion phenotype when it coexisted with F97L mutation (Chua et al., 2003b; Le Pogam et al., 2000). Moreover, it has also been suggested that P5T/A mutation is an independent factor for acute-on-chronic liver failure (ACLF) (Yan et al., 2011; Zhang et al., 2013). However, the mechanism of P5T-associated liver disease progression remains unclear.
To further investigate the potential role of core protein mutations in hepatitis B exacerbation, we cloned the full-length HBV isolates from patients prior to treatment and after emergence of LMV-resistance with exacerbation, and assessed their replication fitness in vitro. Our study revealed that the mutant core, predominantly P5T, boosts the levels of HBV capsid formation and pgRNA encapsidation, and subsequently enhances the viral replication competency, which may contribute to disease progression during LAM treatment. Therefore, the core mutation-mediated enhancement of HBV replication fitness represents an important viral strategy to surmount the antiviral drug pressure due to suppression of polymerase function by NAs.
Materials and Methods
Construction of replication-competent recombinant HBV DNA
Paired serum samples from the patient before treatment (wild type, WT, GenBank Accession No. AY220698; genotype B, serotype adw) and after the lamivudine drug-resistance exacerbation (mutant type, isolate GYF634, GenBank Accession No. AY220697; genotype B, serotype adw) were collected previously (Zhang et al., 2005). The 1.0mer replication-competent HBV genomes were PCR amplified and cloned into vector pUC19 at the SacI restriction site to generate pHBV-WT and pHBV-GYF as previously described (Gunther et al., 1995). To construct plasmid pCMVHBV-WT and pCMVHBV-GYF (1.1 mer, pgRNA transcription is driven by the CMV-IE promoter), the DNA fragment containing HBV nt 1823–3215/1–1822 was retrieved from pHBV-WT and pHBV-GYF by SapI digestion and cloned into the SapI site of plasmid PHY106. pHBV1.3-WT and pHBV1.3-GYF containing HBV nt 957–3215/1–1952 were constructed as previously described (Wang et al., 2007). The core gene point mutations (P5T, S35T, S87G, I97L, Q177K) and the reversion mutations (T5P, T35S, Q79P, D83E, G87S, L97I, K177Q) were individually introduced into pCMVHBV-WT and pCMVHBV-GYF, respectively, by using QuikChange Site-directed Mutagenesis Kit (Stratagene) or Q5 Site-directed Mutagenesis Kit (NEB). The PCR fragment containing the combined 6 core mutations (P5T/S35T/P79Q/E83D/S87G/Q177K) were amplified from pCMVHBV-GYF-L97I and the PCR product was used as primers to mutate pCMVHBV-WT by Q5 mutagenesis, giving rise to plasmid pCMVHBV-WT-6Cmut. The pol-null pCMVHBV-WTΔpol and pCMVHBV-GYFΔpol were generated by mutating the start codon of pol ORF to ACG without changing the amino acid sequence of the overlapping core. The core-null pCMVHBV-WTΔcore and pCMVHBV-GYFΔcore were made through mutating the start codon of core ORF to CTG. The primer sequences for site-directed mutagenesis are listed in Supplemental Table 1. The ORF of core and pol were PCR amplified from pCMVHBV-WT and pCMVHBV-GYF and cloned into pcDNA3 vector to obtain the plasmids expressing wildtype or mutant core and pol. The sequences of plasmids used in this study have been validated by Sanger sequencing.
Drugs
Lamivudine (3TC) was kindly provided by Dr. William Mason (Fox Chase Cancer Center). AT-61 (Delaney et al., 2002) was synthesized by Pharmabridge Inc. GLS-4 (Mani et al., 2018; Wu et al., 2013) and SBA-R01 (Zhou et al., 2017) were kindly provided by Arbutus Biopharma, Inc.
Cell culture and transfection
HepG2 cells and Huh7 cells are maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium (Corning) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 g/ml streptomycin. Cell transfection was conducted with Lipofectamine 2000 (Invitrogen) by following the manufacturer’s instruction.
HBV RNA and DNA analysis
Total cellular RNA was extracted from transfected cells by using the TRIzol reagent (Invitrogen) according to the manufacturer’s specifications. The cytoplasmic encapsidated HBV pgRNA was extracted as previously described (Cai et al., 2016; Liu et al., 2017). HBV RNA Northern blot analysis was performed as previously described (Mao et al., 2013; Mao et al., 2011). HBV core DNA was extracted from transfected cells and subjected to Southern blot analysis as previously described (Cai et al., 2013; Guo et al., 2009; Guo et al., 2007). Membranes were probed with either α−32P-UTP (800 Ci/mmol, Perkin Elmer) labeled plus-strand-specific (for Northern blot hybridization) or minus-strand-specific (for Southern blot hybridization) full-length HBV riboprobe and exposed to a phosphorimager screen. Hybridization signals were quantified with QuantityOne software (Bio-Rad).
HBV particle gel assay
The extracellular HBV particles were analyzed by particle gel assay according to a published protocol (Yan et al., 2017).
The intracellular HBV capsid gel assay was performed as previously described (Yan et al., 2015). Briefly, cells in one well of a 12-well plate were lysed with 200 l of lysis buffer containing 1% NP40. Twenty microliters of the cell lysate were resolved by electrophoresis through a non-denaturing 1% agarose gel, which was then transferred onto the nitrocellulose membrane in TNE buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 1 mM EDTA). HBV capsid and its viral DNA content were detected by enzyme immunoassay (EIA) with a polyclonal anti-core antibody (Dako) and DNA hybridization, respectively, as described in the protocol of particle gel assay (Yan et al., 2017).
Results
Replication fitness of wildtype and LMV-resistant HBV isolates
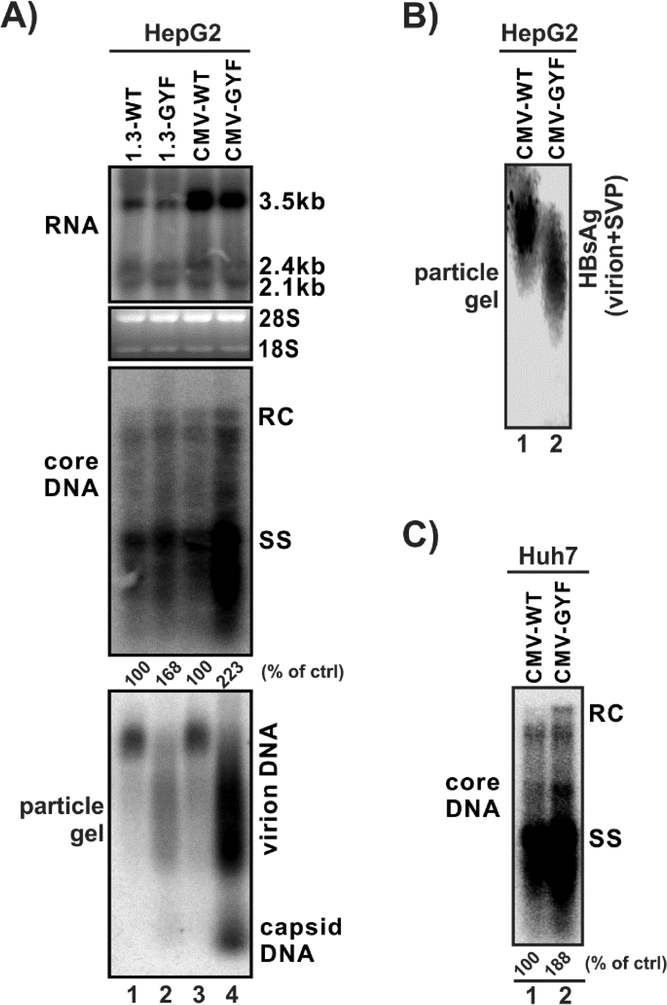
We have previously identified a clinical HBV isolate that exhibited resistance to LMV from a CHB patient undergoing LMV treatment with remarkably high viremia. The patient was initially infected with the wildtype genotype B HBV before the treatment, the drug-resistant mutant virus then became dominant during LMV therapy, resulting in acute exacerbations (Zhang et al., 2005). In the current study, we firstly assessed the replication fitness of this mutant virus, termed GYF, in vitro in HepG2 cell culture. As shown in Fig. 2A, the wildtype (WT) strain and GYF mutant exhibited similar levels of HBV RNA under the transcriptional control of authentic HBV promoters (Fig. 2A, top panel, lanes 1 and 2), indicating that the transcriptional activity of WT and GYF is comparable. When HBV pgRNA is transcribed under the control of CMV-IE promoter, the pgRNA levels are also similar between WT and GYF, though much higher than those transcribed by the HBV core promoter due to stronger activity of the CMV-IE promoter (Fig. 2A, top panel, lanes 3 and 4; and comparing to lanes 1 and 2). However, The GYF isolate exhibited a significantly higher level of intracellular core DNA replication than WT (Fig. 2A, middle panel). In addition, higher levels of HBV virion and DNA-containing naked capsid were found in the supernatant of cells transfected by GYF compared to WT, as revealed by particle gel assay (Fig. 2A, bottom panel). Interestingly, the GYF virions migrate faster than WT in the gel and form a smeared electrophoretic pattern (Fig. 2A, bottom panel), which is possibly due to the mutations on the viral envelope proteins (Zhang et al., 2005) (Fig. 1). Indeed, HBsAg staining of the particle gel demonstrated that the enveloped particles (a mixture of virions (minor species) and subviral particles (major species)) from GYF strain also migrate faster than WT (Fig. 2B). Furthermore, the high DNA replication fitness of GYF was also seen in another hepatoma cell line Huh7 cells (Fig. 2C). Taken together, the high replication fitness of the clinical isolate GYF has been validated in cell cultures, and is mainly attributed to a robust intracellular core DNA replication.
Figure 2. Replication fitness of the wildtype (WT) HBV and the lamivudine-resistant GYF isolate.
(A) HepG2 cells in 12-well-plate were transfected with 1.6 μg of pHBV1.3-WT (lane 1, labeled as 1.3-WT for convenience), pHBV1.3-GYF (lane 2, labeled as 1.3-GYF), pCMVHBV-WT (lane 3, labeled as CMV-WT), pCMVHBV-GYF (lane 4, labeled as CMV-GYF). Cells and supernatant were harvested at day 5 post-transfection for the following analyses: (top panel) Intracellular HBV RNA was detected by Northern blot, the bands of 3.5kb, 2.4/2.1kb HBV RNA are marked. Cellular 28S and 18S ribosomal RNA served as loading control; (middle panel) Cytoplasmic HBV core DNA was determined by Southern blot, the positions of rcDNA (RC) and single-stranded DNA (SS) are labeled; (bottom panel) extracellular HBV virion and naked capsid were analyzed by particle gel assay, the encapsulated viral DNA was detected by hybridization. The relative levels of viral encapsidated pgRNA and core DNA replicative intermediates in each sample are expressed as the percentage of RNA and DNA level in WT samples (lanes 1 and 3), respectively, and indicated underneath the blots. (B) The supernatant samples from panel (A), lanes 3 and 4 were subjected to particle gel assay, and virions and subviral particles (SVP) were detectedby immunoblotting using antibodies against HBsAg. (C) Huh7 cells in 12-well-plate were transfected by CMV-WT or CMV-GYF for 5 days, followed by Southern blot analysis of core DNA. The relative levels of core DNA are expressed as the percentage of that in WT sample.
HBV core protein mutations contribute to the replication enhancement of GYF strain
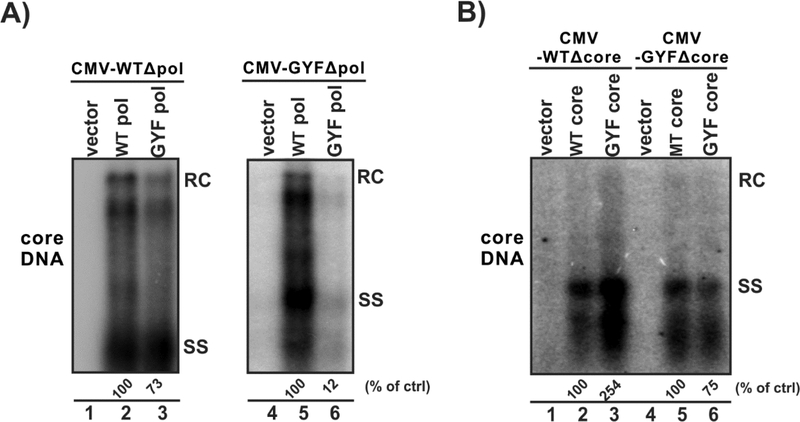
HBV core protein and pol are essential viral factors for core DNA replication. To assess whether the mutations of core or pol play a major role in the elevated replication of GYF, the transcomplementation assay was conducted. First, as shown in Fig. 3A, cotransfection of the pol-null WT (CMV-WTΔpol, left panel) or GYF (CMV-GYFΔpol, right panel) with plasmid expressing WT pol or GYF pol revealed that the mutant pol has less activity to support HBV DNA replication compared to WT pol, which is consistent with previous reports that the NA-resistant pol normally has a reduced reverse transcriptase activity (Zoulim and Locarnini, 2009). Next, core transcomplementation experiment was performed. As shown in Fig. 3B, when core-null WT HBV (CMV-WTΔcore) was cotransfected with GYF core, the viral DNA replication was restored to a significantly higher level than the WT core (lanes 1–3). The same trend was observed in the core-null GYF (CMV-GYFΔcore) group (lanes 4–6). The above results suggest that the core protein of GYF isolate supports a high level of HBV DNA replication, regardless of whether the viral pol is wildtype or LMV-resistant.
Figure 3. Core protein mutations contribute to the enhanced viral replication of GYF isolate.
(A) HepG2 cells in 12-well-plate were transfected with 0.8 μg of pol-null pCMVHBV-WT (CMV-WTΔpol, lanes 1–3) or 0.8 μg of pol-null pCMVHBV-GYF (CMV-GYFΔpol, lanes 4–6) in combination with 0.8 μg control vector (lanes 1 and 4), or 0.8 μg of plasmid expressing WT pol (lanes 2 and 5), or 0.8 μg of plasmid expressing GYF pol (lanes 3 and 6). (B) HepG2 cells were transfected with 0.8 μg of core-null pCMVHBV-WT (CMV-WTΔcore, lanes 1–3) or 0.8 μg of core-null pCMVHBV-GYF (CMV-GYFΔcore, lanes4–6) in combination with 0.8 μg control vector (lanes 1 and 4), or 0.8 μg of plasmid expressing WT core (lanes 2 and 6), or 0.8 μg of plasmid expressing GYF core (lanes 3 and 5). Cells were harvested at day 5 post-transfection and the cytoplasmic HBV core DNA were detected by Southern blot. The relative levels of core DNA are expressed as the percentage of that in the indicated control (ctrl) samples.
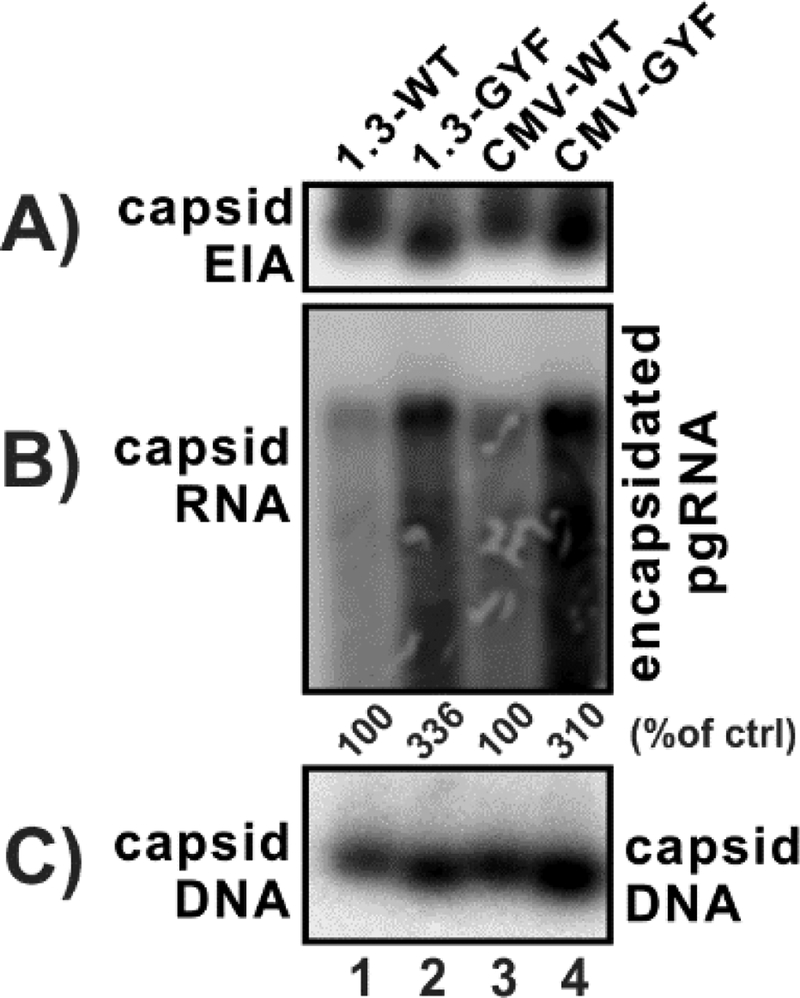
The levels of capsid formation and pgRNA encapsidation of WT and GYF core proteins were then determined. As shown in Fig. 4, GYF core supported a higher level of capsid assembly, and the electrophoretic mobility of GYF capsid was faster than the WT in capsid gel (panel A); more strikingly, the levels of encapsidated pgRNA were significantly higher in GYF strain compared to WT (panel B); as a consequence, and consistent with the results in Fig. 2, GYF strain produced more replicative viral DNA than WT (panel C). Thus, it is inferred that the core mutations in GYF isolate enhance HBV DNA replication primarily through promoting pgRNA encapsidation.
Figure 4. GYF isolate exhibits higher levels of pgRNA encapsidation than WT.
HepG2 cells in 12-well-plate were transfected with 1.6 μg of pHBV1.3-WT (lane 1), pHBV1.3-GYF (lane 2), pCMVHBV-WT (lane 3), pCMVHBV-GYF (lane 4). Cells were harvested at day 5 post-transfection for the following analyses: (A) the cytoplasmic capsid was detected by capsid gel EIA assay using anti-core antibodies; (B) the cytoplasmic encapsidated pgRNA was analyzed by Northern blot, and the relative levels are expressed as the percentage of that in the indicated control samples; (C) the cytoplasmic capsid-containing viral DNA was detected by capsid gel assay through DNA hybridization.
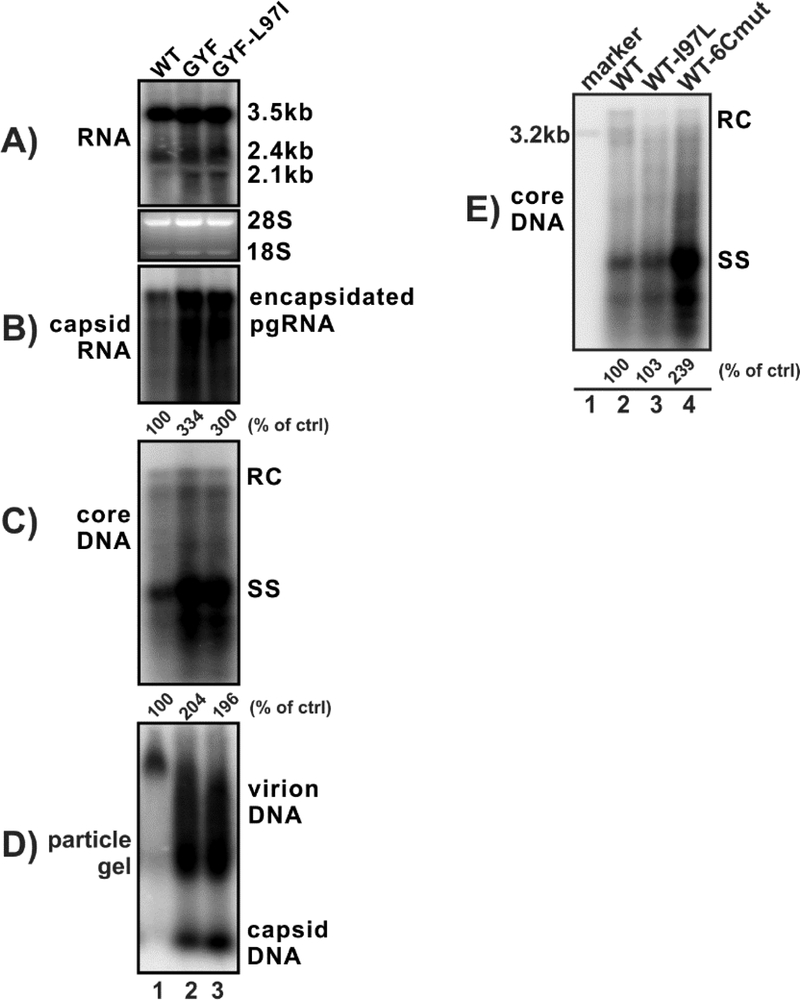
I97L mutation is dispensable for the enhanced viral replication of GYF strain
The mutations within the core protein of GYF mutant include P5T, S35T, P79Q, E83D, S87G, I97L and Q177K. Among those, I97L has been reported to be associated with the increased HBV DNA replication in Huh7 cells (Suk et al., 2002), we thus first examined the potential role of I97L in the replication fitness of GYF isolate. After L97I reversion in the GYF isolate, the levels of pgRNA encapsidation, core DNA replication, and virion secretion, remained unchanged (Fig. 5A–D), indicating that I97L is dispensable for the high replication fitness of GYF isolate. Furthermore, introducing I97L mutation into WT did not enhance viral DNA replication, but changing WT core to other six mutations together (P5T, S35T, P79Q, E83D, S87G, and Q177K) significantly elevated the replication level of WT (Fig. 5E), further suggesting that one or more core mutations other than I97L are responsible for the high replication fitness of GYF isolate.
Figure 5. HBc I97L mutation does not contribute to the elevated replication of GYF strain.
(A-D) HepG2 cells in 12-well-plate were transfected with 1.6 μg of pCMVHBV-WT (lane 1), or pCMVHBV-GYF (lane 2), or pCMVHBV-GYF-L97I (lane 3) for 5 days. The intracellular HBV total RNA (A) and encapsidated pgRNA (B) were analyzed by Northern blot, cytoplasmic core DNA (C) was detected by Southern blot. The extracellular virion and capsid were analyzed by particle gel assay to detect DNA content (D). (E) HepG2 cells in 12-well-plate were transfected with pCMVHBV-WT, or pCMVHBV-WT-L97I, or pCMVHBV-WT-6Cmut for 5 days. HBV core DNA was analyzed by Southern blot. A linear full-length HBV genome DNA serves as 3.2kb size marker. The relative levels of encapsidated pgRNA and core DNA in each sample are expressed as the percentage of RNA and DNA level in the indicated control samples.
P5T promotes HBV replication
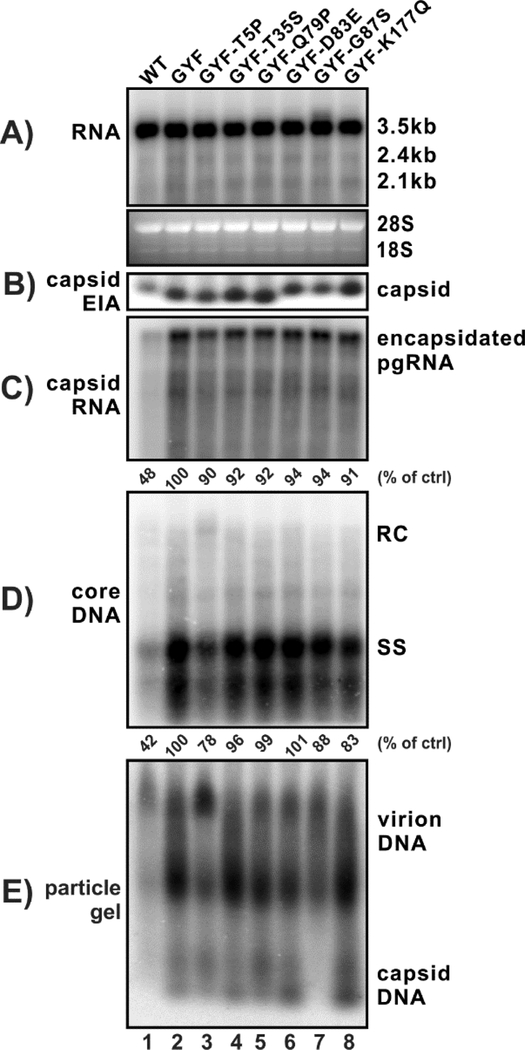
In order to map the functional core mutation(s) responsible for the elevated replication of GYF isolate, we reversed other six core mutations back to the wildtype counterparts individually and assessed their replication in cells. The screening demonstrated that only the T5P restoration in GYF isolate significantly decreased pgRNA encapsidation and core DNA replication (Fig. 6, lane 3 vs 2). The GYF-T5P construct still replicated better than WT (lane 3 vs 1), indicating that other core mutation(s) may also be able to coordinately enhance viral replication (lanes 4–8). Interestingly, the T5P reverse mutation produced slightly more intracellular rcDNA than GYF and restored the wildtype particle gel pattern (lane 3), suggesting that P5 of core may regulate HBV rcDNA synthesis and virion morphogenesis. Furthermore, the G87S reverse mutation eliminated the presence of naked capsid in cell supernatant (lane 7), inferring a potential role of G87 in the secretion of nonenveloped capsid.
Figure 6. The contribution of individual core mutations to the replication capacity of GYF strain.
HepG2 cells were transfected with 1.6 μg of pCMVHBV-WT (lane 1), pCMVHBV-GYF (lane 2), and pCMVHBV-GYF with the indicated single HBcAg amino acid mutation being changed back to its wildtype residue (lane 3–8). Cells were harvested at day 5 posttransfection. Total intracellular viral RNA (A), cytoplasmic capsid (B), encapsidated pgRNA (C), core DNA (D), and supernatant viral particles (E) were analyzed. The relative levels of encapsidated pgRNA and core DNA in each sample are expressed as the percentage of RNA and DNA level in the indicated control samples.
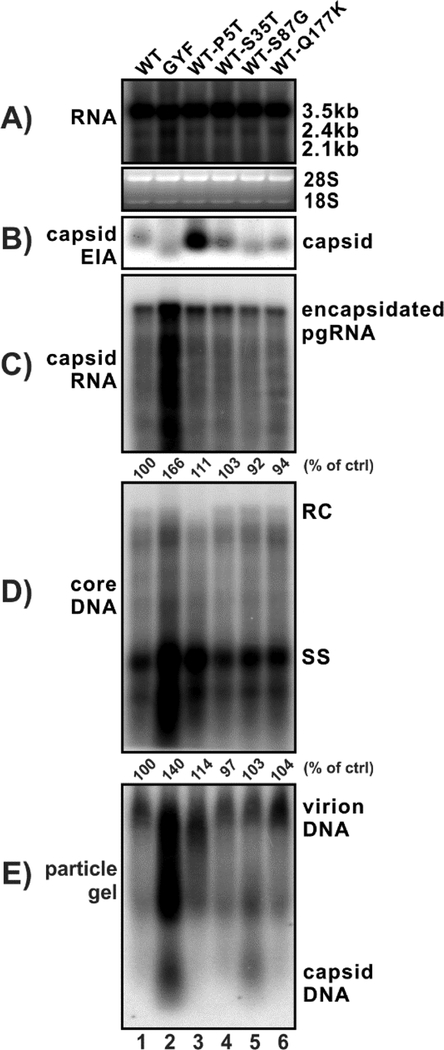
Vice versa, the P5T core mutation of WT HBV markedly upregulated pgRNA encapsidation and ssDNA replication (Fig. 7, lane 3 vs 1). The capsid gel assay demonstrated that P5T mutation exhibited a significant higher EIA signal of capsid (panel B, lane 3), although it remains unknown whether such phenomena results from enhanced capsid formation or a better epitope accessibility of the core antibodies. Furthermore, the P5T single mutation reduced the level of mature rcDNA and resulted in an altered virion electrophoresis pattern (panel D and E, lane 3), which is consistent with the above result (Fig. 6).
Figure 7. The effect of individual core mutants from GYF strain on WT HBV replication.
HepG2 cells were transfected with 1.6 μg of pCMVHBV-WT (lane 1), pCMVHBV-GYF (lane 2), and pCMVHBV-WT with indicated single HBcAg amino acid mutations from the GYF strain (lane 3–6). Cells and supernatant were harvested at day 5 post-transfection for analyses of total intracellular viral RNA (A), cytoplasmic capsid (B), encapsidated pgRNA (C), core DNA (D), and supernatant viral particles (E). The relative levels of encapsidated pgRNA and core DNA in each sample are expressed as the percentage of RNA and DNA level in the indicated control samples.
Other core mutations (S35T, S87G, Q177K) did not obviously affect the replication of WT HBV (lanes 4–6). Considering that the P5T mutant did not completely bring up the WT replication to the level of GYF, it is plausible that the multiple core mutations and/or pol mutations of GYF synergistically contribute to the enhanced pgRNA encapsidation and DNA replication of the GYF strain. Moreover, consistent with the phenotype of G87S reverse mutation shown in Fig. 6, S87G mutation of WT core resulted in an elevated secretion of naked capsid (Fig. 7, lane 5).
Replication of GYF strain is inhibited by CpAMs
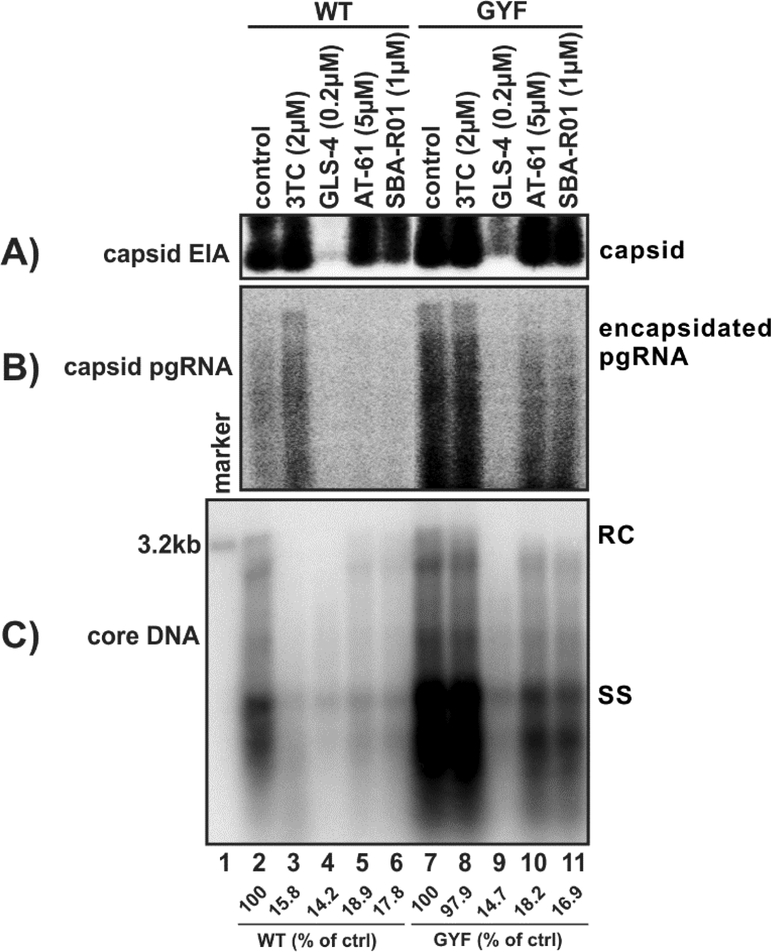
The development of HBV core protein allosteric modulators or capsid assembly modulators (CpAMs) provides a novel opportunity to treat HBV infection through inhibiting viral nucleocapsid formation (Yang and Lu, 2018; Zoulim and Durantel, 2015). Currently, there are three major chemical classes of CpAMs, including heteroaryldihydropyrimidine (HAP) (Deres et al., 2003; Wu et al., 2013), phenylpropenamide (PPA) (Delaney et al., 2002; Feld et al., 2007), and sulfamoylbenzamide (SBA) (Campagna et al., 2013; Lam et al., 2017; Mani et al., 2018). Other chemical scaffolds of CpAMs have also been identified (Corcuera et al., 2018; Lahlali et al., 2018). CpAMs have shown synergistic antiviral effect with NAs, and can be used to treat NA-resistant viruses (Klumpp et al., 2018; Lahlali et al., 2018; Mani et al., 2018). We thus set out to test whether CpAMs could inhibit the GYF strain containing multiple proviral core mutations. As shown in Fig. 8, the replication of WT HBV was significantly inhibited by lamivudine (3TC), the HAP derivative GLS-4, the PPA derivative AT-61, and the SBA reference compound SBA-R01 (lane 2–6). Consistent with the known mechanism of action of these antiviral drugs, 3TC specifically inhibited the reverse transcription of WT HBV and resulted in accumulation of encapsidated pgRNA (lane 3), GLS-4 blocked capsid assembly and reduced the steady state level of core protein (lane 4), AT-61 and SBA-R01 mainly inhibited pgRNA encapsidation without significantly affecting capsid assembly (lanes 5–6). As expected, GYF replicated more efficiently than WT HBV and exhibited resistance to 3TC (lanes 7–8). However, the tested CpAMs significantly inhibited the replication of GYF at the similar efficiencies compared to WT (lanes 9–11). The above results demonstrated that the core mutations of GYF do not confer resistance to CpAMs.
Figure 8. The antiviral effect of NA and CpAMs on WT and GYF strains.
HepG2 cells were transfected with 1.6 μg of pCMVHBV-WT (lanes 2–6) or pCMVHBV-GYF (lanes 7–11). 16 h later, the transfected cells were left untreated (control, lanes 2 and 7) or treated with the indicated compounds at subinhibitory concentrations. The treatment was refreshed every other day, and cells were harvested at day 4 post-treatment. The cytoplasmic capsid (A), encapsidated pgRNA (B), and core DNA (C) were analyzed. The intensities of core DNA signals were quantified and are presented as relative levels compared to the untreated samples (WT or GYF).
Discussion
In clinical practice, the development of HBV drug resistance in CHB patients treated with low barrier NAs, such as lamivudine, is commonly accompanied by virological breakthrough, exacerbation of hepatitis, and even liver failure in severe cases (Zoulim and Locarnini, 2009). The resistance mutations within viral reverse transcriptase region (RT) are selected by NAs, which would however decrease the viral replication capacity (Melegari et al., 1998). Therefore, the underlying mechanism of enhancement of HBV replication and progression of liver disease by certain clinical drug-resistant mutants remains largely unclear. Compensatory mutations with RT regions have been reported to restore the virus replication efficiency under the continuously selection of lamivudine therapy (Ahn et al., 2015; Ghany and Liang, 2007; Ji et al., 2012; Lin et al., 2012; Zoulim and Locarnini, 2009). Our results herein demonstrated that the enhanced nucleocapsid formation and DNA replication of GYF isolate are attributed to core mutations, which represents a previously unknown viral strategy for surmounting antiviral drug pressure.
Our previous study has identified and cloned the full-length HBV isolates from patients prior to treatment and after emergence of LMV-resistance with exacerbation (Zhang et al., 2005). In the present study, we further validated the replication fitness of GYF isolate by systematically analyzing the RNA transcription, capsid assembly, pgRNA encapsidation, DNA replication, and virion secretion in hepatoma cells transiently transfected with the mutant viral genome. Our results demonstrated that GYF isolate produces higher levels of capsid pgRNA and DNA, as well as higher level of virion secretion, than wildtype HBV (Figs. 2 and 4). Furthermore, the trans-complementation experiment revealed that multiple mutations of viral core protein, but not the pol mutants, contribute to the high replication fitness of the GYF isolate (Fig. 3).
Among the 7 concurrent core mutations of GYF isolate, the I97L mutation has been reported to promote HBV DNA replication in cell cultures (Suk et al., 2002). Based on that, we changed I97L mutant back to its wild type sequence within the GYF background. The transient transfection result showed that the isoleucine restoration at codon 97 of core did not alter the capacity of viral DNA replication or virion secretion of GYF isolate (Fig. 5), indicating that I97L mutation does not play a role in enhancing HBV replication, at least not in HepG2 cells. It is worth noting that the previous study indeed observed a much weaker phenotype of I97L-mediated enhancement of HBV replication in HepG2 cells compared to Huh7 cells (Suk et al., 2002).
Further investigation revealed that the mutation from proline (P) to threonine (T) at codon 5 (P5T) of core protein led to enhanced viral replication competency (Figs. 6–7). A previous study reported that the P5T mutation (strain: Shanghai adr) enhanced the intracellular viral DNA replication and rescued the immature secretion phenotype of I97L mutation in Huh7 cells (Chua et al., 2003a). Here, our result demonstrated that P5T alone promoted the viral ssDNA replication but reduced intracellular rcDNA level in HepG2 (Fig. 7); reversely, T5P restoration reduced the DNA replication of GYF isolate but enhanced intracellular rcDNA production (Fig. 6). The slight discrepancy between this study and the previous report may be due to cell type-specific or virus strain-specific effect(s). Moreover, P5T mutation enhanced the EIA signal of capsid in the native gel assay, indicating that P5T may promote capsid assembly and the subsequent pgRNA encapsidation (Fig. 7). Structurally, HBV core protein has a four helix bundle structure, and according to the previously published three-dimensional structure of HBcAg dimer (Klumpp et al., 2015; Packianathan et al., 2010; Wynne et al., 1999), amino acid 5 appears to be located at the interface of two monomers at the N-terminus of HBcAg, indicating that the P5T mutation may regulate the core dimerization/oligomerization to affect nucleocapsid assembly and viral DNA maturation. However, the P5T mutation of WT HBV alone does not completely recapitulate the replication phenotype of GYF isolate, inferring that other core mutation(s) and/or the pol mutation(s) may work together with P5T to further promote virus replication and rcDNA maturation. The synergistic effect among those individual core mutants in the GYF isolate awaits further investigation.
The proline residue at amino acid 5 position of core protein is highly conserved among HBV genotypes. It has been reported that the P5T mutation of HBcAg is common in HBV-related acute-on-chronic liver failure (ACLF) patients (Yan et al., 2011; Zhang et al., 2013). The mechanistic link between P5T mutation and CHB exacerbation including ACLF remains elusive, but may be related to the enhanced virus replication and/or HBcAg-mediated liver inflammation.
Our study suggests that, under circumstances in which the viral polymerase function is suppressed by NAs, enhancement of capsid assembly and viral replication resulting from compensatory core protein mutations is a viable strategy to improve the replication fitness of the virus under NA drug pressure. It is worth noting that core protein mutations are not the classical compensatory mutations which would occur after the emergence of the drug resistance mutations. HBV core mutations could arise in both the wildtype and mutant viruses (Pairan and Bruss, 2009; Tong and Revill, 2016). The mechanism underlying the emergence of compensatory core mutations remain elusive. One limitation of this study is that, though P5T and I97L mutations are commonly found in HBV patients, limited reports are available on the co-existence of multiple core compensatory mutations in NA-resistant patients (Wei et al., 2011; Zhang et al., 2005). The generalizability of our findings should be further evaluated with wider sampling of CHB patients receiving NA therapy.
Nonetheless, the NA-resistant GYF strain harboring compensatory core mutations remains sensitive to non-NA, CpAM type compounds (Fig. 8). This is not unanticipated because none of the core mutations of the GYF isolate overlaps with the reported changes in core protein that reduce sensitivity to CpAMs (Berke et al., 2017; Klumpp et al., 2015; Ruan et al., 2018; Zhou et al., 2017). Thus, CpAMs represent a promising future treatment option for CHB patients, especially those harboring infection with the NA-resistant viruses.
Supplementary Material
Highlights:
Naturally occurring HBcAg mutations compensate for the reduced replication fitness of a lamivudine-resistant HBV variant.
The HBcAg mutations enhance HBV replication primarily through promoting viral nucleocapsid formation.
HBV capsid assembly modulators (CpAMs) efficiently inhibit the mutant virus nucelocapsid formation and DNA replication.
Acknowledgments
We thank Dr. William Mason (Fox Chase Cancer Center), Dr. Xiaodong Xu (Pharmabridge Inc), and the Research Department at Arbutus Biopharma for providing experimental compounds. Dr. Andrea Cuconati is thanked for critical reading of the manuscript. This study is supported by the Natural Science Foundation of Shanghai (18ZR1405600 to YZ), the Research Foundation of Huashan Hospital (North Hospital) (HSBY2016016 to YZ), the National Natural Science Foundation of China (81800529 to YZ, 81672009 to JZ), the Major Science and Technology Special Project of China (2017ZX10202202–001 and 2017ZX10202203-007-002 to JZ), and the US National Institutesof Health grants (AI094474, AI110762, AI123271, and AI134818 to HG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Ahn SH, Kim DH, Lee AR, Kim BK, Park YK, Park ES, Ahn SH, Shin GC, Park S, Kang HS, Rhee JK, Yang SI, Chong Y, Kim KH, 2015. Substitution at rt269 in Hepatitis B Virus Polymerase Is a Compensatory Mutation Associated with Multi-Drug Resistance. PloS one 10, e0136728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL, Brown N, Condreay LD, 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology 27, 1670–1677. [DOI] [PubMed] [Google Scholar]
- Alter H, Block T, Brown N, Brownstein A, Brosgart C, Chang KM, Chen PJ, Chisari FV, Cohen C, El-Serag H, Feld J, Gish R, Glenn J, Greten T, Guo H, Guo JT, Hoshida Y, Hu J, Kowdley KV, Li W, Liang J, Locarnini S, Lok AS, Mason W, McMahon B, Mehta A, Perrillo R, Revill P, Rice CM, Rinaudo J, Schinazi R, Seeger C, Shetty K, Tavis J, Zoulim F, 2018. A research agenda for curing chronic hepatitis B virus infection. Hepatology 67, 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Schaller H, 1992. Hepadnaviral Assembly Is Initiated by Polymerase Binding to the Encapsidation Signal in the Viral-Rna Genome. Embo Journal 11, 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke JM, Tan Y, Verbinnen T, Dehertogh P, Vergauwen K, Vos A, Lenz O, Pauwels F, 2017. Antiviral profiling of the capsid assembly modulator BAY41–4109 on full-length HBV genotype A-H clinical isolates and core site-directed mutants in vitro. Antiviral research 144, 205–215. [DOI] [PubMed] [Google Scholar]
- Birnbaum F, Nassal M, 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol 64, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Guo H, Guo JT, 2007. Molecular virology of hepatitis B virus for clinicians. Clin Liver Dis 11, 685–706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher B, Wynne SA, Crowther RA, 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386, 88–91. [DOI] [PubMed] [Google Scholar]
- Cai D, Nie H, Yan R, Guo JT, Block TM, Guo H, 2013. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods in molecular biology 1030, 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Wang X, Yan R, Mao R, Liu Y, Ji C, Cuconati A, Guo H, 2016. Establishment of an inducible HBV stable cell line that expresses cccDNA-dependent epitope-tagged HBeAg for screening of cccDNA modulators. Antiviral research 132, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT, 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. Journal of virology 87, 6931–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceres P, Stray SJ, Zlotnick A, 2004. Hepatitis B virus capsid assembly is enhanced by naturally occurring mutation F97L. J Virol 78, 9538–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PK, Wen YM, Shih C, 2003a. Coexistence of two distinct secretion mutations (P5T and I97L) in hepatitis B virus core produces a wild-type pattern of secretion. Journal of virology 77, 7673–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PK, Wen YM, Shih C, 2003b. Coexistence of Two Distinct Secretion Mutations (P5T and I97L) in Hepatitis B Virus Core Produces a Wild-Type Pattern of Secretion. Journal of Virology 77, 7673–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcuera A, Stolle K, Hillmer S, Seitz S, Lee JY, Bartenschlager R, Birkmann A, Urban A, 2018. Novel non-heteroarylpyrimidine (HAP) capsid assembly modifiers have a different mode of action from HAPs in vitro. Antiviral research. [DOI] [PubMed] [Google Scholar]
- Cui X, Luckenbaugh L, Bruss V, Hu J, 2015. Alteration of Mature Nucleocapsid and Enhancement of Covalently Closed Circular DNA Formation by Hepatitis B Virus Core Mutants Defective in Complete-Virion Formation. Journal of virology 89, 10064–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney W.E.t., Edwards R, Colledge D, Shaw T, Furman P, Painter G, Locarnini S, 2002. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrobial agents and chemotherapy 46, 3057–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney W.E.t., Yang H, Westland CE, Das K, Arnold E, Gibbs CS, Miller MD, Xiong S, 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol 77, 11833–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H, 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299, 893–896. [DOI] [PubMed] [Google Scholar]
- Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA, 2007. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral research 76, 168–177. [DOI] [PubMed] [Google Scholar]
- Ghany M, Liang TJ, 2007. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology 132, 1574–1585. [DOI] [PubMed] [Google Scholar]
- Gish R, Jia JD, Locarnini S, Zoulim F, 2012. Selection of chronic hepatitis B therapy with high barrier to resistance. The Lancet. Infectious diseases 12, 341–353. [DOI] [PubMed] [Google Scholar]
- Gunther S, Li BC, Miska S, Kruger DH, Meisel H, Will H, 1995. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol 69, 5437–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Jiang D, Ma D, Chang J, Dougherty AM, Cuconati A, Block TM, Guo JT, 2009. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. Journal of virology 83, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT, 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. Journal of virology 81, 12472–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu K, 2017. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D, Liu Y, Li L, Xu Z, Si LL, Dai JZ, Li X, Wang L, Yao Z, Xin SJ, Chen GF, Xu D, 2012. The rtL229 substitutions in the reverse transcriptase region of hepatitis B virus (HBV) polymerase are potentially associated with lamivudine resistance as a compensatory mutation. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 54, 66–72. [DOI] [PubMed] [Google Scholar]
- Klumpp K, Lam AM, Lukacs C, Vogel R, Ren S, Espiritu C, Baydo R, Atkins K, Abendroth J, Liao G, Efimov A, Hartman G, Flores OA, 2015. High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proceedings of the National Academy of Sciences of the United States of America 112, 15196–15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp K, Shimada T, Allweiss L, Volz T, Lutgehetmann M, Hartman G, Flores OA, Lam AM, Dandri M, 2018. Efficacy of NVR 3–778, Alone and In Combination With Pegylated Interferon, vs Entecavir In uPA/SCID Mice With Humanized Livers and HBV Infection. Gastroenterology 154, 652–662e658. [DOI] [PubMed] [Google Scholar]
- Lahlali T, Berke JM, Vergauwen K, Foca A, Vandyck K, Pauwels F, Zoulim F, Durantel D, 2018. Novel potent capsid assembly modulators regulate multiple steps of the Hepatitis B virus life-cycle. Antimicrobial agents and chemotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AM, Ren S, Espiritu C, Kelly M, Lau V, Zheng L, Hartman GD, Flores OA, Klumpp K, 2017. Hepatitis B Virus Capsid Assembly Modulators, but Not Nucleoside Analogs, Inhibit the Production of Extracellular Pregenomic RNA and Spliced RNA Variants. Antimicrobial agents and chemotherapy 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampertico P, Liaw YF, 2012. New perspectives in the therapy of chronic hepatitis B. Gut 61 Suppl 1, i18–24. [DOI] [PubMed] [Google Scholar]
- Le Pogam S, Yuan TT, Sahu GK, Chatterjee S, Shih C, 2000. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J Virol 74, 9099–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Chien RN, Hu CC, Lai MW, Yeh CT, 2012. Identification of hepatitis B virus rtS117F substitution as a compensatory mutation for rtM204I during lamivudine therapy. J Antimicrob Chemother 67, 39–48. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan R, Guo JT, Block TM, Mechti N, Guo H, 2017. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS pathogens 13, e1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani N, Cole AG, Phelps JR, Ardzinski A, Cobarrubias KD, Cuconati A, Dorsey BD, Evangelista E, Fan K, Guo F, Guo H, Guo JT, Harasym TO, Kadhim S, Kultgen SG, Lee ACH, Li AHL, Long Q, Majeski SA, Mao R, McClintock KD, Reid SP, Rijnbrand R, Snead NM, Micolochick Steuer HM, Stever K, Tang S, Wang X, Zhao Q, Sofia MJ, 2018. Preclinical Profile of AB-423, an Inhibitor of Hepatitis B Virus Pregenomic RNA Encapsidation. Antimicrobial agents and chemotherapy 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo JT, Guo H, 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS pathogens 9, e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Zhang J, Jiang D, Cai D, Levy JM, Cuconati A, Block TM, Guo JT, Guo H, 2011. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. Journal of virology 85, 1048–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melegari M, Scaglioni PP, Wands JR, 1998. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology 27, 628–633. [DOI] [PubMed] [Google Scholar]
- Ning X, Luckenbaugh L, Liu K, Bruss V, Sureau C, Hu J, 2018. Common and Distinct Capsid and Surface Protein Requirements for Secretion of Complete and Genome-Free Hepatitis B Virions. Journal of virology 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono SK, Kato N, Shiratori Y, Kato J, Goto T, Schinazi RF, Carrilho FJ, Omata M, 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Invest 107, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packianathan C, Katen SP, Dann CE 3rd, Zlotnick A, 2010. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. Journal of virology 84, 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairan A, Bruss V, 2009. Functional surfaces of the hepatitis B virus capsid. Journal of virology 83, 11616–11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoud C, Seigneres B, Wang Z, Trepo C, Zoulim F, 1999. Transient selection of a hepatitis B virus polymerase gene mutant associated with a decreased replication capacity and famciclovir resistance. Hepatology 29, 230–237. [DOI] [PubMed] [Google Scholar]
- Ruan L, Hadden JA, Zlotnick A, 2018. Assembly properties of Hepatitis B Virus core protein mutants correlate with their resistance to assembly-directed antivirals. Journal of virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon J, Rodes B, Zoulim F, Bartholomeusz A, Soriano V, 2006. Mutations affecting the replication capacity of the hepatitis B virus. J Viral Hepat 13, 427–434. [DOI] [PubMed] [Google Scholar]
- Suk FM, Lin MH, Newman M, Pan S, Chen SH, Liu JD, Shih C, 2002. Replication advantage and host factor-independent phenotypes attributable to a common naturally occurring capsid mutation (I97L) in human hepatitis B virus. Journal of virology 76, 12069–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Mason WS, 1982. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell 29, 403–415. [DOI] [PubMed] [Google Scholar]
- Tong S, Revill P, 2016. Overview of hepatitis B viral replication and genetic variability. Journal of hepatology 64, S4–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Xu X, Luo C, Ma ZM, Jiang HL, Ding JP, Wen YM, 2007. Mutational analysis revealed that conservation of hepatitis B virus reverse transcriptase residue 306 (rtP306) is crucial for encapsidation of pregenomic RNA. FEBS Lett 581, 558–564. [DOI] [PubMed] [Google Scholar]
- Wei C, Chong YT, Wen JZ, Li YW, Li G, 2011. Characterization of hepatitis virus B isolated from a multi-drug refractory patient. Virus research 155, 254–258. [DOI] [PubMed] [Google Scholar]
- Wu G, Liu B, Zhang Y, Li J, Arzumanyan A, Clayton MM, Schinazi RF, Wang Z, Goldmann S, Ren Q, Zhang F, Feitelson MA, 2013. Preclinical characterization of GLS4, an inhibitor of hepatitis B virus core particle assembly. Antimicrobial agents and chemotherapy 57, 5344–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG, 1999. The crystal structure of the human hepatitis B virus capsid. Molecular cell 3, 771–780. [DOI] [PubMed] [Google Scholar]
- Yan R, Cai D, Liu Y, Guo H, 2017. Detection of Hepatitis B Virus Particles Released from Cultured Cells by Particle Gel Assay. Methods in molecular biology 1540, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhao X, Cai D, Liu Y, Block TM, Guo JT, Guo H, 2015. The Interferon-Inducible Protein Tetherin Inhibits Hepatitis B Virus Virion Secretion. Journal of virology 89, 9200–9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Li K, Li F, Su H, Mu J, Tong S, Patel M, Xia J, Wands JR, Wang H, 2011. T1846 and A/G1913 are associated with acute on chronic liver failure in patients infected with hepatitis B virus genotypes B and C. Journal of medical virology 83, 996–1004. [DOI] [PubMed] [Google Scholar]
- Yang L, Lu M, 2018. Small Molecule Inhibitors of Hepatitis B Virus Nucleocapsid Assembly: A New Approach to Treat Chronic HBV Infection. Curr Med Chem 25, 802–813. [DOI] [PubMed] [Google Scholar]
- Yuan TT, Sahu GK, Whitehead WE, Greenberg R, Shih C, 1999a. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J Virol 73, 5731–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TT, Tai PC, Shih C, 1999b. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human hepatitis B virus core antigen. J Virol 73, 10122–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wan Z, You S, Liu H, Zhu B, Chen J, Rong Y, Zang H, Li C, Wang H, Xin S, 2013. Association of Hepatitis B Virus Mutations of A1846T and C1913A/G With Acute-on-Chronic Liver Failure Development From Different Underlying Chronic Liver Diseases. Hepat Mon 13, e12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Yao X, Wang YX, Liu F, Ma ZM, Weng XH, Wen YM, 2005. High replicative full-length lamivudine-resistant hepatitis B virus isolated during acute exacerbations. Journal of medical virology 77, 203–208. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hu T, Zhou X, Wildum S, Garcia-Alcalde F, Xu Z, Wu D, Mao Y, Tian X, Zhou Y, Shen F, Zhang Z, Tang G, Najera I, Yang G, Shen HC, Young JA, Qin N, 2017. Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms. Scientific reports 7, 42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Durantel D, 2015. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harbor perspectives in medicine 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Locarnini S, 2009. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 137, 1593–1608e1591–1592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.