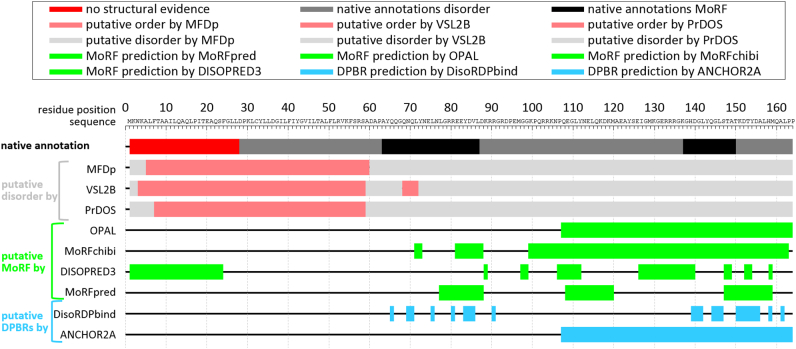
Fig. 2.
Prediction of IDRs, MoRFs and disordered protein-binding regions (DPBRs) for the T-cell surface glycoprotein CD3 zeta chain (UniProt id: P20963). The native annotations are shown using horizontal line immediately below the protein sequence at the top of the figure where regions without structural evidence are in red, IDRs are in grey and MoRFs are in black. The following three lines show putative annotations of disorder produced with three leading predictors: MFDp, VSL2B and PrDOS where putative IDRs are in grey and putative structured regions in rose. The next four lines give the putative MoRFs generated with MoRFpred, OPAL, MoRFchibi and DISOPRED3 (in green). The two lines at the bottom correspond to putative DBPRs predicted with DisoRDPbind and ANCHOR2A (in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)