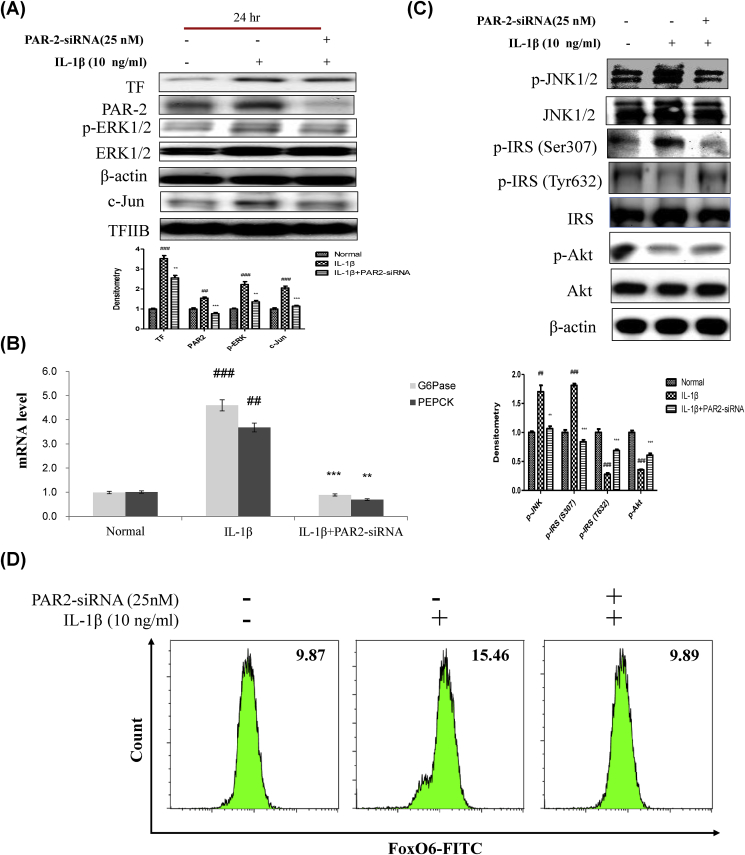
Fig. 6.
IL-1β induced insulin resistance via the PAR2 pathway. (A) HepG2 cells were grown to 80% confluence in 100-mm dishes in DMEM medium, pre-treated (1 day) with or without PAR2-siRNA (25 nM), and then stimulated with IL-1β (10 ng/ml) and analyzed by Western blotting using antibodies. Bars in densitometry data represent means ± SE, and significance was determined using an unpaired t-test: ##p < 0.01, ###p < 0.001 vs. Normal; ∗∗p < 0.01, ∗∗∗p < 0.001 vs. IL-1β treated cells. (B) Gluconeogenesis genes (G6P and PEPCK) were subjected to real-time qRT-PCR analysis. Results of one factor ANOVA: ##p < 0.01, ###p < 0.001 vs. Normal; ∗∗p < 0.01, ∗∗∗p < 0.001 vs. IL-1β-treated group. (C) HepG2 cells were transiently transfected pre-incubated with PAR2-siRNA (25 nM) for 24 h with or without IL-1β (10 ng/ml). Cells were analyzed by Western blotting using p-JNK1/2, JNK1/2, p-IRS (S307), p-IRS (T632), IRS, p-Akt, and Akt antibodies. Bars in densitometry data represent means ± SE, and significance was determined using an unpaired t-test: ##p < 0.01, ###p < 0.001 vs. Normal; ∗∗p < 0.01, ∗∗∗p < 0.001 vs. IL-1β treated cells. (D) HepG2 cells were transiently transfected with PAR2-siRNA or control siRNA for 24 h using lipofectamine 2000 and then stimulated with IL-1β (10 ng/ml) for 24 h. The cells were stained with anti-FoxO6 and FITC goat anti-Rabbit IgG.