Abstract
Nutrition influences both hepatic function and aging, but mechanisms are poorly understood. Here, the effects of lifelong, ad libitum-fed diets varying in macronutrients and energy on hepatic gene expression were studied. Gene expression was measured using Affymetrix mouse arrays in livers of 46 mice aged 15 months fed one of 25 diets varying in protein, carbohydrates, fat, and energy density from 3 weeks of age. Gene expression was almost entirely influenced by protein intake. Carbohydrate and fat intake had few effects on gene expression compared with protein. Pathways and processes associated with protein intake included those involved with mitochondrial function, metabolic signaling (PI3K-Akt, AMPK, mTOR) and metabolism of protein and amino acids. Protein intake had variable effects on genes associated with regulation of longevity and influenced by caloric restriction. Among the genes of interest with expression that were significantly associated with protein intake are Cth, Gls2, Igf1, and Nnmt, which were increased with higher protein intake, and Igf2bp2, Fgf21, Prkab2, and Mtor, which were increased with lower protein intake. Dietary protein has a powerful impact on hepatic gene expression in older mice, with some overlap with genes previously reported to be involved with regulation of longevity or caloric restriction.
Keywords: Geometric Framework, Liver, Nutrition, Life span
Nutrition has profound effects on aging. The major nutritional intervention that has been studied is caloric restriction, whereby lifelong reduction in energy intake by 10%–50% without malnutrition has been reported to increase life span and delay biological changes of aging in many species (1,2). There has been increasing interest in discovering which genes and pathways mediate this response, partly because this can provide a platform for the discovery of pharmacological agents that delay aging (3). Key nutrient-sensing pathways that are thought to regulate aging include mTOR (mechanistic target of rapamycin), SIRT1 (sirtuin 1), insulin/IGF-1 (insulin-like growth factor 1), AMPK (AMP-activated protein kinase), and more recently FGF21 (fibroblast growth factor 21) (4–6). The effects of caloric restriction on mammalian gene expression have also been assessed to provide data-driven insights into pathways and genes that influence healthy aging (7,8). In one meta-analysis of more than 50 animal studies, it was found that caloric restriction is associated with overexpression of 101 genes and underexpression of another 73 genes. Pathways affected by caloric restriction included growth hormone signaling, lipid metabolism, and immune responses (8). Another similar meta-analysis identified genes associated with oxidative stress, inflammation, and tumorigenesis (7).
Although caloric restriction has been a foundation of aging research for nearly a century, more recent studies have questioned this model (1). The effects of caloric restriction on aging have been shown to vary by species, strain, and sex (9), and although reduced energy intake is a key contributor to its benefits (10,11), other factors such as periodic fasting (12) and reduced intake of macronutrients, especially protein (5), may also be important. Recent studies in insects and mice have indicated that the ratio of macronutrients in the diet can also influence life span, with most studies reporting that diets composed of low protein combined with high carbohydrates increase life span in ad libitum-fed animals (13–15). Nutrient composition has been found to influence aging in various other ad libitum-fed animal models of aging including monkeys (16) and mice (17). From a translational perspective, this is important because restricted feeding is not sustainable for most humans in developed countries. In terms of mechanisms, studies to date suggest that low-protein, high-carbohydrate diets decrease mTOR activity while reducing circulating IGF-1 and increasing FGF-21 levels (6, 14).
As yet, there have not been any reports of the effects of low-protein, high-carbohydrate diets on gene expression, nor the long-term effects of dietary macronutrients on gene expression. The liver is the primary organ that responds to nutrition and regulates systemic metabolic responses to diet, and hepatic gene expression is therefore highly responsive to dietary perturbations. There have been many, mostly short-term studies of the effects of diet on the hepatic transcriptome that have identified a wide range of genes and pathways that are influenced by nutrition and these are mostly involved with the regulation of the metabolism of nutrients and energy substrates (18).
Here, we studied gene expression in the livers of old mice maintained on diets varying systematically in protein, carbohydrate, fat, and energy content and compared the outcomes with published data on gene expression in aging and caloric restriction. We used the Geometric Framework for Nutrition (GFN) in the design of both the dietary treatment and the analytical approach because this facilitates interpretation of the effects of all macronutrients and their interactions, rather than the effects of each nutritional parameter evaluated individually (19,20).
Methods
Livers were analyzed from a mouse study previously published (14). Briefly, 3-week-old male and female mice (C57Bl6/J, n = 858, Animal Resources Centre, WA, Australia) were ad libitum-fed one of 25 experimental diets varying in protein, carbohydrate, fat, and energy content (Specialty Feeds, WA, Australia). Energy manipulations were achieved by addition of cellulose generating low-, medium-, and high-energy-density diets (8, 13, and 17 kJ/g, respectively). At 15 months of age, one cohort of mice spanning the diets was euthanized and tissues collected, whereas the remaining mice were maintained for life-span analysis. All protocols were performed in accordance with the Sydney Local Health District Animal Welfare Committee (protocol no. 2009/003).
Frozen liver tissue samples from mice at 15 months of age were sectioned into 10-mg blocks, and DNA, RNA, and protein were extracted using the Qiagen AllPrep Mini Kit. RNA was isolated (TRI reagent, Sigma-Aldrich, NSW, Australia) and analyzed using Affymetrix Mouse Gene ST arrays at the Ramaciotti Centre for Genomics, University of New South Wales (Sydney, NSW, Australia). The results of gene expression in these liver samples have been reported in a previous publication (GEO: GSE85998) (6). The sex, diets, and dietary intakes of the mice are shown in Supplementary Table 1.
In addition, liver samples that had undergone microarray assays were run on quantitative polymerase chain reaction (PCR) plates using the RT2 profiler array using the Mouse Insulin Signaling Pathway kit (Qiagen, catalogue no. PAMM-030Z). A pooled sample was serially diluted as a standard curve in the Fluidigm Biomark software for PCR analysis.
Because the gene that was most positively correlated with protein intake was Cth (cystathionine gamma-lyase), hydrogen sulfide production capacity was measured in freeze–thaw homogenates of liver using the lead sulfide method as described previously (21).
Data analyses were based on macronutrient intake (kilojoule per mouse per day). The relationship between macronutrients and gene expression was determined using two methods. First, the correlation between the intake of each macronutrient and the expression of each gene was calculated using a Pearson’s correlation coefficient. Second, the Geometric Framework approach was used where the statistical significance of the relationships between each gene and macronutrients and the interactions between macronutrients was calculated with Generalized Additive Models (GAMS). In addition to the GAMS statistics, graphical representation of the relationship between macronutrients and gene expression was assessed using colored surfaces as described previously (14). All calculations were performed in R (Version 3.4.0, The R Foundation for Statistical Computing) or Excel 2013.
Genes of interest were determined from the p values returned by each of these methods and application of Benjamini–Hochberg correction with a false discovery rate of .05. Heatmaps were performed in R using the gplots package. Gene enrichment analysis was performed in EnrichR to determine gene pathways based on the KEGG database and biological pathways based on the GO Biological Processes 2017 (22). Only pathways and processes with more than two genes represented and a false discovery rate of <.05 were considered to be significant.
Results
The Effects of Macronutrients on Overall Liver Gene Expression
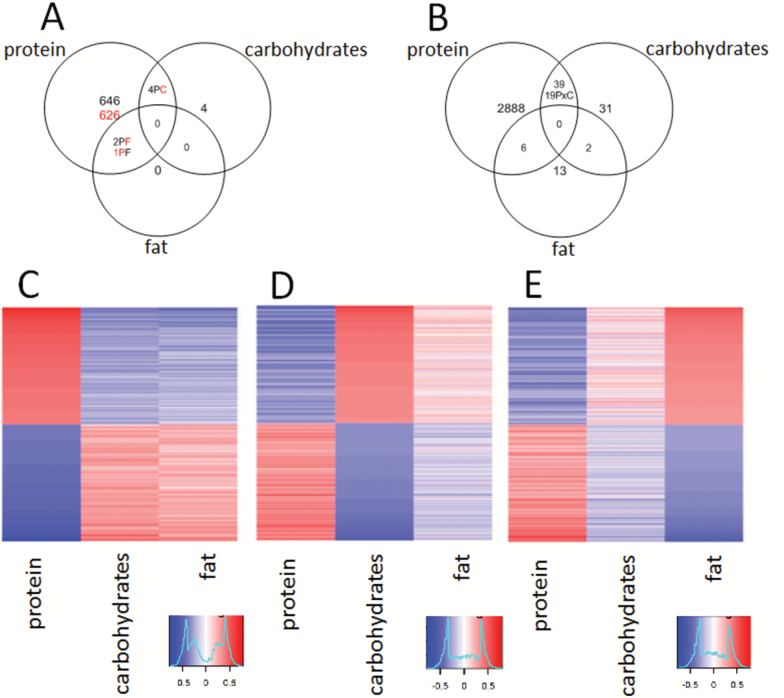
The most highly expressed gene was Alb (albumin), and there was about a 100-fold difference in the expression of the highest versus the lowest expressed genes within each liver. Of the three macronutrients, dietary protein was associated with changes in the expression of the highest number of genes using either correlation analysis (n = 1,279 genes, 648 positive correlation, 631 negative correlation) or GAMS (n = 2,933 genes). This compared to only 8 genes by correlation analysis and 72 genes by GAMS for dietary carbohydrates, and only 3 genes by correlation analysis and 19 genes by GAMS for dietary fat (Figure 1A and B). For dietary protein, there were 980 genes (77% of genes from correlation analysis) in common using either GAMS or correlation analysis, whereas for dietary carbohydrates, there were 4 genes (50% of genes from correlation analysis) in common using both types of analysis, indicating that both types of analysis are identifying similar overall trends.
Figure 1.
(A) Venn diagrams showing the effects of macronutrient intake on gene expression in the liver. The results determined using Pearson’s correlation coefficient (black = positive correlation, red = negative correlation). (B) The results determined using GAMS (P × C = interactive term between carbohydrates and protein). Heatmaps comparing the overall pattern of expression of genes in the liver. The values are Pearson’s correlation coefficients ranked from most positive correlation in red to the most negative correlation in blue. The most positively correlated 1,000 genes and the most negatively correlated 1,000 genes for each macronutrient are included. The genes have been ranked according to the significance of the correlation with protein (C), carbohydrates (D), and fat (E). The heatmap for energy is shown in Supplementary Figure 5.
Heatmap analysis (Figure 1C–E) revealed a very strong inverse relationship between gene expression that correlated with protein intake compared with intake of fat or carbohydrates, such that genes that were positively correlated with the intake of dietary protein were negatively correlated with the intake of either fat or carbohydrates, and vice versa. Although the inverse relationships between the macronutrients on the heatmaps are very distinct, it should be noted that only a few genes were statistically significantly associated with the intakes of carbohydrates or fats. Fat and carbohydrate intake are the main contributors to total energy intake; therefore, the heatmap for total energy intake mirrored that seen for carbohydrates and fat (Supplementary Figure 5).
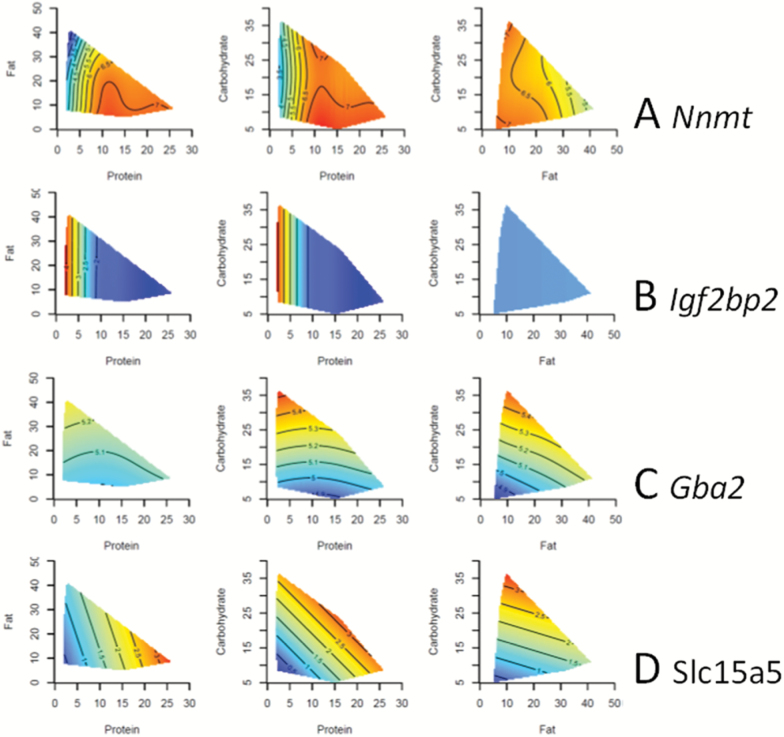
The genes with the most statistically significant association with macronutrient intake are shown in Table 1. Four representative surfaces using the Geometric Framework approach are shown in Figure 2. These genes are shown because they were highly significantly associated with macronutrient intake (Supplementary Table 2) and demonstrate characteristic responses to macronutrients that can be easily visualized by the Geometric Framework method. Nicotinamide N-methyltransferase (Nnmt) was positively associated with protein intake (Figure 2A), whereas Insulin-like growth factor 2 mRNA-binding protein 2 (Igf2bp2) was negatively associated with protein intake (Figure 2B). Glucosylceramidase beta 2 (Gba2) was positively associated with carbohydrate intake (Figure 2C), whereas Solute carrier family 15 member 5 (Slc15a5) was positively associated with both protein and carbohydrate intake (Figure 2D).
Table 1.
Gene Expression With the Highest Statistical Association With Macronutrient Intake Determined Using Either GAMS or Correlation Analysis
| Protein | p | Carbohydrates | p | Fat | p |
|---|---|---|---|---|---|
| Top genes by GAMS analysis | |||||
| Igf2bp2 | 7.34E−21 | Slc15a5 | 5.83E−07 | Sag | 2.9E−11 |
| Adgrg2 | 6.31E−12 | Reg1 | 3.11E−06 | Gm5424 | 9.18E−09 |
| Orm1 | 1.71E−10 | Sox13 | .00072 | Hsd3b1 | 2.58E−07 |
| Nnmt | 1.57E−10 | Alb | .00073 | Cps1 | 1.99E−05 |
| Igsf23 | 6.3E−10 | Lrrc16a | .00108 | Cfap54 | .00054 |
| Spink5 | 6.72E−10 | Fam131c | .00168 | Mir192 | .00150 |
| Gm5424 | 6.24E−09 | Cyp4a12a | .00220 | Gpc1 | .00288 |
| Corin | 1.63E−08 | Hsd17b6 | .00358 | X1700012C14Rik | .00759 |
| Itga6 | 9.14E−08 | Rnf128 | .00610 | Zfp449 | .01249 |
| Impa2 | 1.83E−07 | Gba2 | .00927 | Fam19a5 | .01529 |
| Gpi1 | 2.41E−07 | Rtn4 | .01255 | Pcdh18 | .02078 |
| Slc15a5 | 2.57E−07 | Elovl3 | .01706 | Gfod2 | .02631 |
| Fgf21 | 3.06E−07 | X1700012C14Rik | .01613 | Sema5b | .02444 |
| Vtcn1 | 2.95E−07 | Wasf3 | .01623 | Sult1e1 | .02970 |
| Psat1 | 3.09E−07 | Pdilt | .01556 | Rnf133 | .03283 |
| Arhgef2 | 9.22E−07 | Adgrg7 | .01509 | Trpc3 | .03193 |
| Hsd17b6 | 1.05E−06 | Derl3 | .01469 | Slc39a12 | .03276 |
| Slc13a2 | 1.09E−06 | Pard3b | .01428 | Sel1l3 | .03158 |
| Cps1 | 1.16E−06 | Cyp2u1 | .01425 | Cidec | .03688 |
| Soat2 | 5.73E−06 | Hsd3b5 | .01567 | ||
| Top genes by correlation analysis, positive correlation with intake | |||||
| Cth | 1.92E−05 | Slc17a8 | .00711 | Gpc1 | .038492 |
| Lrtm1 | 2.64E−05 | Pard3b | .01925 | ||
| Slc13a2 | 2.36E−05 | Gba2 | .02124 | ||
| Gpx6 | 6.05E−05 | Tubg1 | .02399 | ||
| Acmsd | 9.08E−05 | A230050P20Rik | .02411 | ||
| Sdhb | .00022 | Fam131c | .02692 | ||
| Nnmt | .00021 | Unc13b | .02818 | ||
| Acadsb | .00022 | Ergic1 | .03640 | ||
| Gm5424 | .00025 | ||||
| Slc43a1 | .00037 | ||||
| Top genes by correlation analysis, negative correlation with intake | |||||
| Adgrg2 | 4.78E−06 | Cps1 | .00722 | ||
| Impa2 | 2.72E−05 | Gm5424 | .01158 | ||
| Igf2bp2 | 4.72E−05 | ||||
| Slc9a7 | 4.76E−05 | ||||
| Unc13b | 5.69E−05 | ||||
| Lhx6 | 6.77E−05 | ||||
| Slc17a8 | .00025 | ||||
| App | .00028 | ||||
| Slc7a7 | .00030 | ||||
| Gpi1 | .00032 | ||||
Note: For correlation analysis, genes are divided into those with positive and negative correlation. Benjamini–Hochberg correction with a false discovery rate of .05 has been performed for the p values. Genes in the top 20 for both GAMS and correlation analysis are bolded.
Figure 2.
Three-dimensional response surfaces created using the Geometric Framework for Nutrition. The relationships between the macronutrients and four genes [(A) Nnmt, (B) Igf2bp2, (C) Gba2, (D) Slc15a5] are demonstrated. The response surfaces vary from red, which is the highest value, to blue, which is the lowest value. The axes show the food intake in kilojoule per mouse per day. Each of the three surfaces shows the interaction between two macronutrients at the median value of the other macronutrient. The GAMS statistics are shown in Supplementary Table 2.
The Effects of Macronutrients on Gene Pathways
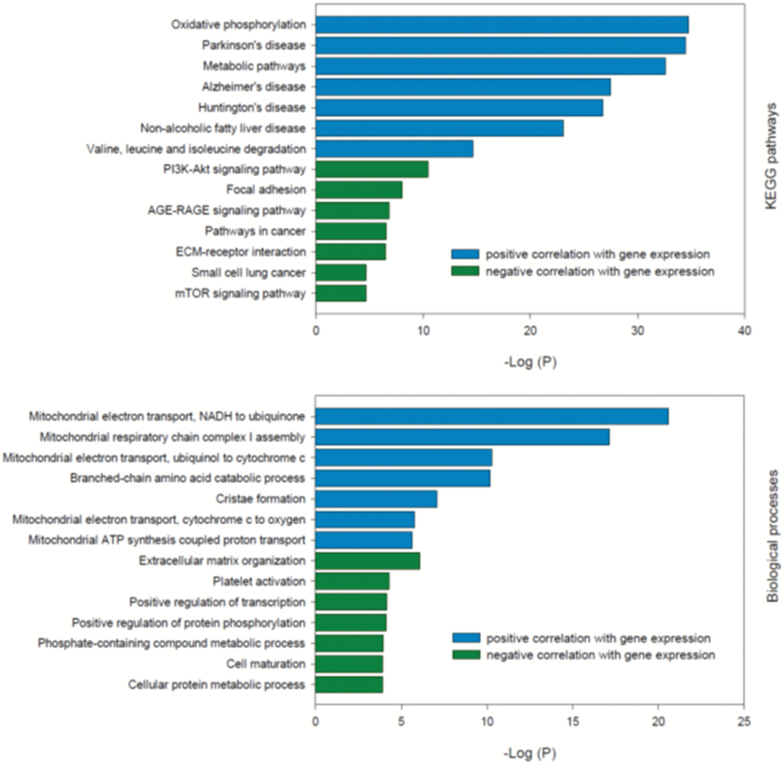
Gene pathway analyses based on correlation coefficients were analyzed according to whether the genes had a positive or a negative correlation with macronutrient intake (Supplementary Table 3, Figure 3A). Pathways enriched by genes whose expression was positively correlated with protein intake included oxidative phosphorylation and multiple pathways associated with amino acid metabolism, as well as pathways associated with neurodegenerative diseases. Pathways enriched by genes whose expression was negatively correlated with protein intake include several key metabolic signaling pathways (PI3K-Akt, mTOR, AMPK) and insulin signaling pathways, as well as pathways associated with various cancers. Gene pathway analysis based on GAMS only found five pathways associated with protein intake, all of which were also present in the pathways found using correlation analysis. There were few if any significant or relevant pathways linked to carbohydrate or fat intake with either analytical approach.
Figure 3.
Gene pathways and biological processes associated with protein intake, where the association between protein intake and gene expression has been determined using Pearson’s correlation coefficient. Pathways analysis was performed using Enrichr. Full datasets are provided in Supplementary Tables 3 and 4.
The Effects of Macronutrients on Biological Processes
Biological processes associated with genes whose expression was positively correlated with protein intake included a very large number of processes associated with mitochondria as well as amino acid metabolism (Supplementary Table 4, Figure 3B). Biological processes associated with genes whose expression was negatively correlated with protein intake included transcription, protein metabolism, and cell differentiation and maturation. There were no pathways associated with fat intake and only two with carbohydrate intake, and none identified using GAMS analysis.
The Effect of Macronutrients on Longevity Regulating and Aging Pathways
The KEGG “longevity regulating pathways, multiple species” gene set (ko0413, http://www.genome.jp/dbget-bin/www_bget?pathway+ko04213) includes 59 genes of which 50 were present in this microarray data set, some with two variants. Using GAMS analysis, there were only seven genes in common with these longevity regulating genes (Atg5, Mtor, Pik3r2, Igf1r, Prkab2, Hsap1b, Adcy3) and three using correlation analysis (Atg4, Mtor, Pik3r2). The correlation coefficients between each of these genes and the intake of macronutrients and total energy were plotted on a heatmap (Supplementary Figure 1) to determine the overall pattern of the relationship between macronutrient intake and expression of genes associated with the regulation of aging. There were many more genes whose expression were negatively correlated with protein intake than positively correlated with protein intake. Genes associated with carbohydrate intake had the opposite pattern. Consequently, there was a clear inverse pattern between genes associated with protein intake versus those associated with carbohydrate intake (and a similar, but less marked pattern when comparing protein intake and fat intake). Fat and carbohydrate intake are the main contributors to total energy intake; therefore, the heatmap for total energy intake mirrored that seen for carbohydrates and fat.
The GenAge database (http://genomics.senescence.info/genes/) includes 43 antilongevity genes and 81 prolongevity genes that were also present in this microarray dataset. As with the Kegg database, there was an inverse relationship between protein versus carbohydrates, fat, and total energy (Supplementary Figure 2).
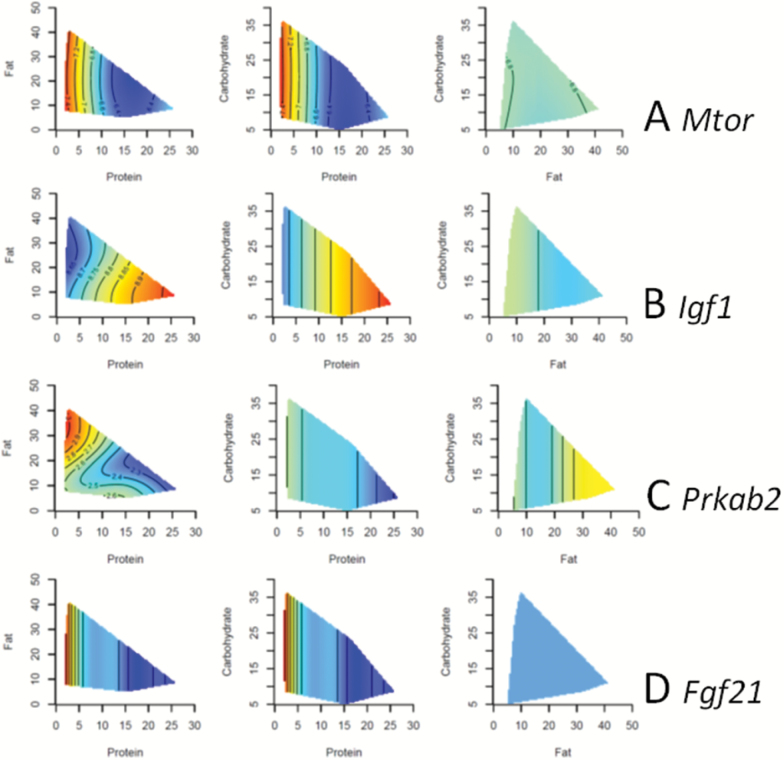
The Geometric Framework was used to evaluate five nutrient-sensing genes that are considered to link nutrition with aging: Mtor, Igf1, Sirt1, Prkab2 (a subunit of AMPK), and Ffg21 (Figure 4, Supplementary Table 5) (5,6). Surprisingly, the expression of Mtor, which is one of the key signaling pathways stimulated by protein, was upregulated with lower protein intake. We have previously published the effects of macronutrients on mTOR protein phosphorylation in these mice (14), and comparison of these data with Mtor gene expression showed that there was no correlation (r = −.052, p = .7). Low protein intake was associated with reduced expression of Igf1 and increased expression of Prkab2 and Fgf21. The expression of Sirt1 was not significantly influenced by dietary macronutrients in this study.
Figure 4.
Three-dimensional response surfaces created using the Geometric Framework for Nutrition. The relationships between the macronutrients and four nutrient-sensing genes [(A) Mtor, (B) Igf1, (C) Pkrab2, (D) Fgf21] are demonstrated. The response surfaces vary from red, which is the highest value, to blue, which is the lowest value. The axes show the food intake in kilojoule per mouse per day. Each of the three surfaces shows the interaction between two macronutrients at the median value of the other macronutrient. The GAMS statistics are shown in Supplementary Table 5.
The genes influenced by protein intake were compared with those reported to be influenced by caloric restriction. A meta-analysis identified 174 genes where expression is influenced by caloric restriction (8), of which only 30 were in common with those genes influenced by protein intake determined by GAMS and only 11 in common with those genes influenced by protein intake determined by correlation (Supplementary Table 6).
PCR Confirmation of Hepatic Microarray Analysis
To validate the results of the transcriptome analysis, quantitative reverse transcription PCR was performed on a set of 84 genes associated with nutrient-sensing growth pathways. The expression patterns of the majority of genes from the microarray were closely correlated with their PCR counterparts (Supplementary Tables 7–8).
Hydrogen Sulfide Production by the Liver
The gene with expression that was most positively correlated with protein intake was Cth, which is a key gene involved in the production of hydrogen sulfide gas by liver. Therefore, we measured hydrogen sulfide production by the liver from another 52 mice that spanned all the diets. Hydrogen sulfide production and Cth expression were closely related, both increased by high protein intake and decreased by low protein intake (Supplementary Figure 3, Supplementary Table 9).
Discussion
In a review of 172 publications on the effects of nutrients on the hepatic transcriptome, Osada (18) reported that protein intake influenced genes involved with lipogenesis, fatty acid uptake, oxidative stress, and DNA methylation; carbohydrate intake influenced genes involved with oxidative stress, cell proliferation, ammonium, Pparga, and Fxr; and fat intake influenced inflammation, beta-oxidation, and several genes including A2m, Slc13a5, Nrep, Cyp3a, and Scd1. The studies in the review were mostly short term over days and weeks and used heterogeneous dietary interventions. By contrast, our long-term study over 15 months examining the effects of 25 different diets varying in macronutrients showed that protein intake was the most powerful driver of gene expression, whereas the intake of carbohydrates and fat had limited effects that were in the opposite direction to changes seen with protein intake. Fat and carbohydrate intake are the main contributors to total energy intake; therefore, the responses to total energy intake reflected those seen for carbohydrates and fat. It appears that with aging and/or very long-term dietary exposure, only protein intake remains a potent regulator of hepatic gene expression, which presumably reflects a greater biological imperative to regulate long-term protein metabolism by the liver. It is possible that muscle and adipose tissue play a greater role in responding to long-term changes in dietary carbohydrate and fat, but we did not measure gene expression in these tissues.
There have been a few other studies that have examined the effects of differing amounts of macronutrients on hepatic gene expression over longer-term periods, and these reported similar effects to those we observed with dietary protein. Diaz-Rua and colleagues (23) fed rats two diets differing in protein content (protein:carbohydrate:fat 20:70:10 vs. 45:45:10) and determined effects on hepatic transcriptome after 4 months. They identified 30 genes that were the most relevant genes affected by a high-protein diet in the liver, of which 14 were also identified in our study (Slc7a2, Slc43a1, Agxt, Got1, Gpt, Hpd, Sds, Asl, Cps1, Nnmt, Slc4a2, Kcnma1, Pgam2, Dgat1). By comparison, Schwarz and colleagues (24) fed mice one of three diets (protein:carbohydrate:fat 15:75:10, 15:50:35, or 50:15:35) for 12 weeks and then measured hepatic gene expression. They identified 154 genes where expression was increased by dietary protein, and these were involved with amino acid and nitrogen metabolism or energy and oxidative metabolism and included 12 of these same genes identified by Diaz-Rua and colleagues (23). Gene pathway analysis (using Enrichr and the KEGG database) showed that these genes are involved with the biosynthesis of amino acids and the metabolism of alanine, aspartate, and glutamate, whereas a biological process analysis (using Enrichr and the GO Biological Process 2017) showed that they are involved with the urea cycle and amino acid biosynthetic processes. These pathways and biological processes have obvious implications for the metabolism of dietary protein. It should be noted that the quality (ie, source) of dietary protein has also been reported to influence hepatic transcriptome (25,26).
We have previously shown that low-protein, high-carbohydrate diets are associated with longer life span in ad libitum-fed mice, a finding that is supported by numerous insect studies (14,15). Most previous studies on nutrition and aging have shown that caloric restriction delays aging and increases health span (1,2,27,28), and there have been many studies investigating the effects of caloric restriction on gene expression (8,29). Therefore, we compared gene expression in our study with published databases on genes whose expression is influenced by aging or caloric restriction across species and tissues. When compared with the KEGG longevity regulating pathways, we only found 7 out of 59 genes in common (Atg5, Mtor, Pik3r2, Igf1r, Prkab2, Hsap1b, Adcy3), which suggests that low-protein, high-carbohydrate diets might influence life span by alternative pathways, although this could be secondary to methodological issues between studies. However, it should be noted that these seven genes include pathways that are critical to the response of aging to nutrients including autophagy, mTOR, IGF-1, and AMPK. We also compared gene expression in our study with published data on caloric restriction from a meta-analysis (8) and only found a few genes in common (6%–17% depending on analytic method). This lack of overlap once again suggests that low-protein, high-carbohydrate diets influence aging via different pathways. Therefore, we interrogated our data for pathways that are generally believed to be crucial for mediating the effects of nutrition on aging: mTOR, AMPK, SIRT1, IGF-1, and FGF21 (Figure 4). Low protein intake was associated with increased Ffg21 and Prkab2 expression and reduced Igf-1 expression, which reflect the direction of change seen with caloric restriction. We have previously shown that circulating FGF21 and IGF-1 are similarly influenced by macronutrients in these same mice (6). However, surprisingly Mtor expression was increased with low protein intake, despite our previous observation that mTOR phosphorylation was decreased with low-protein, high-carbohydrate diets (14). We did not find any correlation between phospho-mTOR and Mtor expression, so this disparity probably reflects the differences between transcription, translation, and activation. Overall, it appears that the effects of macronutrients and their interactions on gene expression in the liver do not substantially overlap with effects that have previously been found to be associated with caloric restriction or regulation of longevity; however, there are some key pathways that are concordant including IGF-1, AMPK, and FGF21. In a very recent study of short-term caloric restriction on the hepatic transcriptome of mice, Derous and colleagues (30) found that the expression of IFG-1, mTOR, and nuclear factor kappa beta were altered in a manner consistent with increased life span, but not the sirtuins or FGF21. It should be noted that a high-protein ad libitum-fed diet was associated with reduced energy intake through protein leverage (31); therefore, a high protein intake may induce some changes in gene expression similar to those seen in caloric restriction because of concomitant reduction in calorie intake.
We also found many individual genes of interest because of their statistical association with macronutrients and their involvement metabolic responses and aging, and some of these are discussed below. For example, Velazquez-Villegas and colleagues found that high ratios of dietary protein to carbohydrates (protein:carbohydrate:fat 50:33:17 vs. 20:63:17 vs. 5:77:18) fed to mice more than 8 days increased the hepatic expression of glutaminase 2 (Gls2) (32). Likewise, Schwarz and colleagues found that high-protein diets increased expression of Gls2 in mice after 1 and 12 weeks (24). Gls2 is the main glutaminase expressed in the liver and increases protein catabolism via the urea cycle. Gls2 has been reported to be upregulated in response to high-protein diets, inflammation, and diabetes (32). In our study, Gls2 was also positively correlated with protein intake according to both GAMS and correlation analysis and was among the top 20 genes influenced by protein intake (Table 1, Supplementary Figure 4, Supplementary Table 10). Analysis by the GFN showed that Gls2 expression was mostly influenced by protein intake, with a small effect of fat intake and no effect of carbohydrate intake. Of course it is not unexpected for a gene to be involved in protein catabolism to be upregulated in response to high dietary protein intake, and our data show that this response is maintained long term and into old age.
Garcia Caraballo and colleagues (33) studied the effects of seven diets varying in macronutrients (protein 11%–58%, carbohydrates 0%–81%, fat 8%–42%) for a period of 3 weeks on hepatic gene expression. They found that high-protein, low-carbohydrate diets reduced Fgf21 and Pparg expression, whereas using the GFN we found that these were only reduced by protein intake and not influenced by the other macronutrients. They also found that Acaca (acetyl-CoA carboxylase 1) was influenced by the ratio of carbohydrates to fat. On GFN analysis, we also found that the interactive term between carbohydrates and fat was associated with Acaca expression and protein (Supplementary Figure 4, Supplementary Table 10). The overlapping results between the two studies indicate that macronutrients individually and interactively can influence the expression of genes involved in fatty acid and glucose metabolism, as well as the key metabolic hormone, FGF21. Unlike Garcia Caraballo, we did not observe any changes in the expression of two other key metabolic regulators, Pck1 or Fasn.
The gene for cystathionine gamma-lyase, Cth, had the strongest positive correlation with protein intake, whereas GAMS analysis showed that it was not influenced by the other macronutrients (Supplementary Figure 3, Supplementary Table 9). This gene was also identified by Schwarz and colleagues as being upregulated with high-protein diets (24) and is important in the liver for the production of hydrogen sulfide (transsulfuration pathway). Deletion of Cth is associated with reduced inflammatory and injury responses to sepsis (34), whereas hydrogen sulfide and Cth have been shown to be a key mechanism for the benefits of short-term dietary restriction (21). Hydrogen sulfide production by the liver increases in response to caloric restriction and may represent a stress resistance response (9). We found that the expression of Cth paralleled the production of hydrogen sulfide in the liver (Supplementary Figure 3). This raises the possibility that Cth might be an important mechanism linking protein intake with aging. However, it should be noted that Hine and colleagues found that short-term dietary restriction of sulfur amino acids increased the expression of Cth and mediated stress resistance induced by caloric restriction (21). On the other hand, in this study we found that long-term high-protein diets were associated with increased Cth expression and reduced life span. The transsulfuration pathway undergoes different changes in the liver compared with other tissues in old age (35), and our experiments were long term in old mice, whereas those of Hine and colleagues were short term in young mice. It is also possible that upregulation of the Cth pathway may be part of a compensatory response to protect against the negative aging effects of a high-protein diet.
There are limitations to our study. Only a subset of 46 mice of the total of 183 euthanized at 15 months of age were used for gene expression studies, although they did span the full range of diets. Only a single tissue and single strain of mouse were analyzed, and the data were not robust enough to study the effects of sex. There is no established statistical method to evaluate gene expression across a large range of diets, so we used two methods: correlation analysis with intake of each macronutrient and a GAMS analysis where all the macronutrients and their interactions are assessed within a single statistical model. The advantage of correlation analysis is that the direction of change is immediately apparent. However the use of GAMS and GFN is preferable because it allows interactive terms to be assessed (36). Encouragingly both methods generated similar conclusions in terms of the predominant effect of protein on gene expression, and the pathways, processes, and genes that were identified. This adds additional robustness to our conclusions.
In summary, protein was the strongest driver of gene expression in the livers of old mice. Protein intake was associated with pathways involved with energy and amino acid metabolism. Although there was only some overlap with genes known to be associated with regulation of longevity and caloric restriction, protein intake was associated with changes in several key nutrient-sensing pathways that influence aging including AMPK, mTOR, IGF-1, and FGF21.
Funding
The study was supported by the Australian National Health and Medical Research Council (Project grants 571328, 1084267, and 1101913 to D.L.C., S.J.S., D.R.), NHMRC Peter Doherty Biomedical Fellowship (SSB), the Ageing and Alzheimers Institute, and the Sydney Medical School Foundation.
Conflict of Interest
None reported.
Supplementary Material
References
- 1. Ingram DK, de Cabo R. Calorie restriction in rodents: caveats to consider. Ageing Res Rev. 2017;39:15–28. doi:10.1016/j.arr.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mercken EM, Carboneau BA, Krzysik-Walker SM, de Cabo R. Of mice and men: the benefits of caloric restriction, exercise, and mimetics. Ageing Res Rev. 2012;11:390–398. doi:10.1016/j.arr.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le Couteur DG, McLachlan AJ, Quinn RJ, Simpson SJ, de Cabo R. Aging biology and novel targets for drug discovery. J Gerontol A Biol Sci Med Sci. 2012;67:169–174. doi:10.1093/gerona/glr095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. doi:10.1038/nrg2188 [DOI] [PubMed] [Google Scholar]
- 5. Solon-Biet SM, Mitchell SJ, de Cabo R, Raubenheimer D, Le Couteur DG, Simpson SJ. Macronutrients and caloric intake in health and longevity. J Endocrinol. 2015;226:R17–R28. doi:10.1530/JOE-15-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solon-Biet SM, Cogger VC, Pulpitel T, et al. . Defining the nutritional and metabolic context of FGF21 using the geometric framework. Cell Metab. 2016;24:555–565. doi:10.1016/j.cmet.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 7. Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi:10.1186/1471-2164-10-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plank M, Wuttke D, van Dam S, Clarke SA, de Magalhães JP. A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Mol Biosyst. 2012;8:1339–1349. doi:10.1039/c2mb05255e [DOI] [PubMed] [Google Scholar]
- 9. Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, et al. . Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 2016;23:1093–1112. doi:10.1016/j.cmet.2016.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swindell WR. Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev. 2012;11:254–270. doi:10.1016/j.arr.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Speakman JR, Mitchell SE, Mazidi M. Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp Gerontol. 2016;86:28–38. doi:10.1016/j.exger.2016.03.011 [DOI] [PubMed] [Google Scholar]
- 12. Lee C, Longo VD. Fasting vs dietary restriction in cellular protection and cancer treatment: from model organisms to patients. Oncogene. 2011;30:3305–3316. doi:10.1038/onc.2011.91 [DOI] [PubMed] [Google Scholar]
- 13. Lee KP, Simpson SJ, Clissold FJ, et al. . Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc Natl Acad Sci USA. 2008;105:2498–2503. doi:10.1073/pnas.0710787105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solon-Biet SM, McMahon AC, Ballard JW, et al. . The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi:10.1016/j.cmet.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Couteur DG, Solon-Biet S, Cogger VC, et al. . The impact of low-protein high-carbohydrate diets on aging and lifespan. Cell Mol Life Sci. 2016;73:1237–1252. doi:10.1007/s00018-015-2120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mattison JA, Colman RJ, Beasley TM, et al. . Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8:14063. doi:10.1038/ncomms14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–B501. [DOI] [PubMed] [Google Scholar]
- 18. Osada J. The use of transcriptomics to unveil the role of nutrients in Mammalian liver. ISRN Nutr. 2013;2013:403792. doi:10.5402/2013/403792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simpson SJ, Le Couteur DG, Raubenheimer D. Putting the balance back in diet. Cell. 2015;161:18–23. doi:10.1016/j.cell.2015.02.033 [DOI] [PubMed] [Google Scholar]
- 20. Simpson SJ, Raubenheimer D. Caloric restriction and aging revisited: the need for a geometric analysis of the nutritional bases of aging. J Gerontol A Biol Sci Med Sci. 2007;62:707–713. [DOI] [PubMed] [Google Scholar]
- 21. Hine C, Harputlugil E, Zhang Y, et al. . Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160:132–144. doi:10.1016/j.cell.2014.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuleshov MV, Jones MR, Rouillard AD, et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi:10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Díaz-Rúa R, Keijer J, Palou A, van Schothorst EM, Oliver P. Long-term intake of a high-protein diet increases liver triacylglycerol deposition pathways and hepatic signs of injury in rats. J Nutr Biochem. 2017;46:39–48. doi:10.1016/j.jnutbio.2017.04.008 [DOI] [PubMed] [Google Scholar]
- 24. Schwarz J, Tomé D, Baars A, Hooiveld GJ, Müller M. Dietary protein affects gene expression and prevents lipid accumulation in the liver in mice. PLoS One. 2012;7:e47303. doi:10.1371/journal.pone.0047303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Endo Y, Fu Z, Abe K, Arai S, Kato H. Dietary protein quantity and quality affect rat hepatic gene expression. J Nutr. 2002;132:3632–3637. doi:10.1093/jn/132.12.3632 [DOI] [PubMed] [Google Scholar]
- 26. Song S, Hooiveld GJ, Li M, et al. . Distinct physiological, plasma amino acid, and liver transcriptome responses to purified dietary beef, chicken, fish, and pork proteins in young rats. Mol Nutr Food Res. 2016;60:1199–1205. doi:10.1002/mnfr.201500789 [DOI] [PubMed] [Google Scholar]
- 27. McCay C, Crowell M, Maynard L. The effect of retarded growth upon the length of life and upon ultimate size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- 28. Masoro EJ, Shimokawa I, Yu BP. Retardation of the aging processes in rats by food restriction. Ann NY Acad Sci. 1991;621:337–352. [DOI] [PubMed] [Google Scholar]
- 29. Swindell WR. Comparative analysis of microarray data identifies common responses to caloric restriction among mouse tissues. Mech Ageing Dev. 2008;129:138–153. doi:10.1016/j.mad.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Derous D, Mitchell SE, Wang L, et al. . The effects of graded levels of calorie restriction: XI. Evaluation of the main hypotheses underpinning the life extension effects of CR using the hepatic transcriptome. Aging (Albany NY). 2017;9:1770–1824. doi:10.18632/aging.101269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simpson SJ, Raubenheimer D. Obesity: the protein leverage hypothesis. Obes Rev. 2005;6:133–142. doi:10.1111/j.1467-789X.2005.00178.x [DOI] [PubMed] [Google Scholar]
- 32. Velázquez-Villegas LA, Charabati T, Contreras AV, Alemán G, Torres N, Tovar AR. PPARα downregulates hepatic glutaminase expression in mice fed diets with different protein: carbohydrate ratios. J Nutr. 2016;146:1634–1640. doi:10.3945/jn.116.232868 [DOI] [PubMed] [Google Scholar]
- 33. Garcia Caraballo SC, Comhair TM, Dejong CHC, Lamers WH, Koehler SE. Dietary treatment of fatty liver: high dietary protein content has an antisteatotic and antiobesogenic effect in mice. Biochim Biophys Acta. 2017;1863:1789–1804. doi:10.1016/j.bbadis.2017.04.022 [DOI] [PubMed] [Google Scholar]
- 34. Gaddam RR, Fraser R, Badiei A, et al. . Cystathionine-gamma-lyase gene deletion protects mice against inflammation and liver sieve injury following polymicrobial sepsis. PLoS One. 2016;11:e0160521. doi:10.1371/journal.pone.0160521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Predmore BL, Alendy MJ, Ahmed KI, Leeuwenburgh C, Julian D. The hydrogen sulfide signaling system: changes during aging and the benefits of caloric restriction. Age (Dordr). 2010;32:467–481. doi:10.1007/s11357-010-9150-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson SJ, Raubenheimer D. Perspective: tricks of the trade. Nature. 2014;508:S66. doi:10.1038/508S66a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.