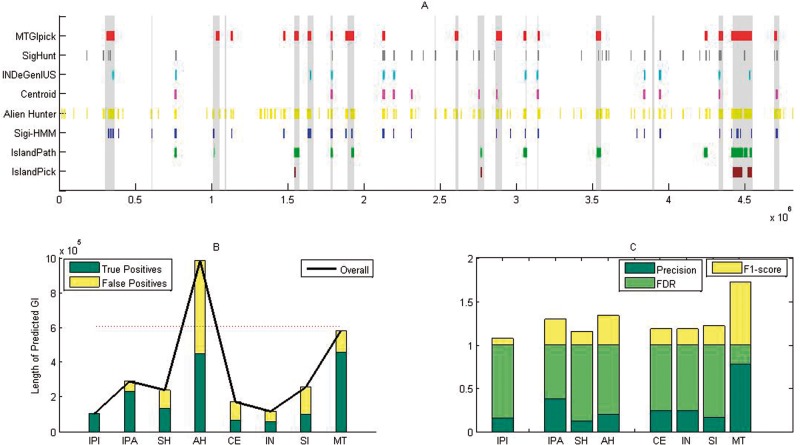
Figure 4.
Performance of the proposed MTGIpick (MT), SIGI-HMM (SH), Alien_Hunter (AH), centroid (CE), IslandPath-DIMOB (IPA), INDeGenIUS (IN), SigHunt (SI) and IslandPick (IPI) on the detection of genomic islands in S. enterica serovar typhi CT18. (A) Predicted GIs found by all of the methods, and the known genomic islands are shown as vertical bars. (B) Overall length of the predicted genomic islands, true positives and false positives of all of the evaluated methods at the nucleotide level. (C) Precision, false discovery rate (FDR) and F1 score of all of the evaluated methods at the island level, in which the precision, false-positive rate and F1 score are calculated based on the number of known GIs, which are covered by > 50% of the results of the prediction method.