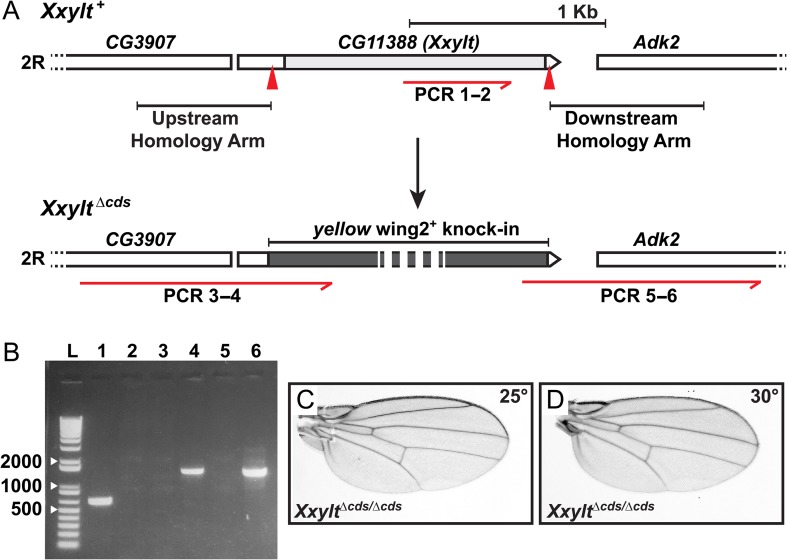
Fig. 2.
Loss of Xxylt does not show impaired Notch signaling phenotypes in flies. (A) Generation of a CG11388 loss-of-function allele using CRISPR/Cas9 engineering. Top panel shows schematic representation of the genomic region of second chromosome (2 R) containing CG11388 (Xxylt) and its neighboring genes. Light gray box indicates the coding sequence. Red arrowheads mark the sgRNA binding sites. Homology arms used for homologous recombination-mediated repair are shown upstream and downstream to the sgRNA binding sites. The red half arrow indicates a region of the CG11388 coding sequence used to confirm deletion by PCR. Bottom panel shows yellow wing2+ knock-in (dark gray box) and extended upstream and downstream homology regions used to confirm the knock-in. Numbers written under the red half arrows refer to the lanes on the PCR gel shown in panel B. (B) PCR amplification products for CG11388 internal (lanes 1 and 2), extended upstream homology region (lanes 3 and 4) and extended downstream homology region (lanes 5 and 6) from y w (control; lanes 1, 3 and 5) and XxyltΔcds/Δcds (lanes 2, 4 and 6) genomic DNA. Absence of PCR amplification product for CG11388 internal and presence of extended upstream and downstream homology regions confirmed the replacement of CG11388 with yellow wing2+. L marks the DNA ladder. (C and D) Wings from adult XxyltΔcds/Δcds flies raised at 25°C (C) and 30°C (D) show no sign of impaired Notch signaling.