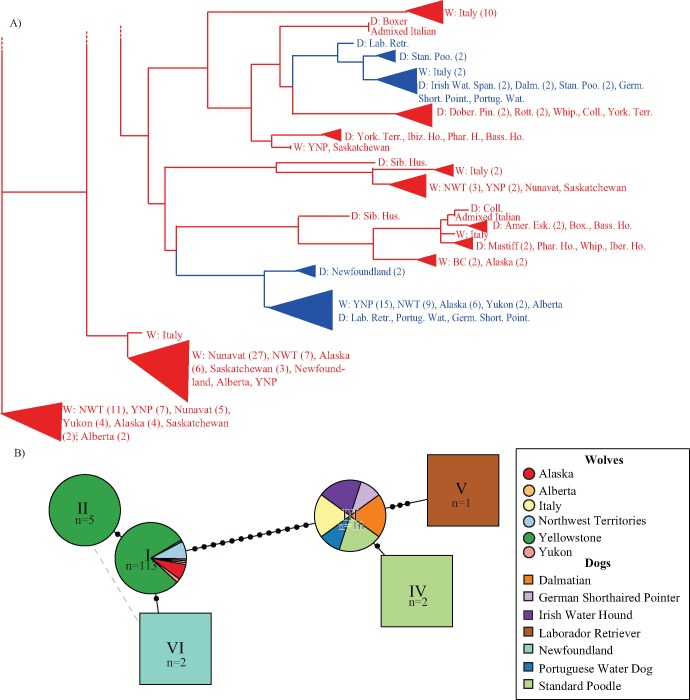
Fig. 7.
Neighbor joining tree and haplotype network of the K locus suggest a single introgression event. (A) Partial neighbor-joining tree of 26 kb core nonrecombining region, based on pairwise nucleotide distances between haplotypes from 172 unrelated individuals. Haplotypes are labeled as belonging to a dog breed (“D”) or wolf population (“W”) and colored according to KB (blue) or ky (red). All nodes had 100% support from 5,000 bootstrap replicates. (B) Haplotype network of all KB haplotypes sampled. Each haplotype (n = 134) consists of a 11,754 bp phased haplotype representing sites that passed filters within the 26 kb nonrecombining region around the K locus deletion. Seven unique KB haplotypes are represented by roman numerals, with the number of haplotypes within each type indicated. Nodes are colored and shaped according to geographic origin and species (see key). Haplotype I includes wolves from Alaska (n = 8), Alberta (n = 1), Northwest Territories (n = 9), Yellowstone (n = 90), and Yukon (n = 2), and dogs (n = 1 for each of German short-haired pointer, labrador retriever, and Portuguese water dog). Haplotype IV includes Dalmatians (n = 2), German Shorthaired Pointers (n = 1), Irish Water Spaniels (n = 2), Portuguese Water Dogs (n = 1), Standard Poodles (n = 2), and Italian wolves (n = 2).