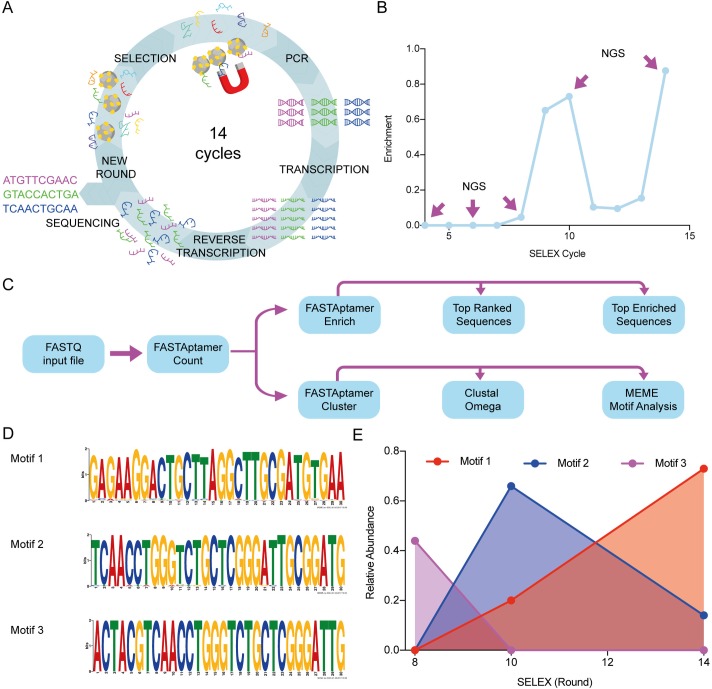
Fig 2. NGS allows the identification of aptamer motifs and their evolution during SELEX.
(A) Workflow of the SELEX technique and variations applied in this work. The ssDNA library (N40) was incubated with the HMG-box proteins immobilized in magnetic beads. After incubation, a magnet is used to separate the ssDNA molecules that bound the proteins on the magnetic beads. To amplify the bound molecules a PCR using a modified reverse primer with the T7 promoter region is performed. After purification, in vitro transcription is carried out, followed by in vitro reverse transcription. Finally, an enriched ssDNA library is generated and used for a next cycle or for sequencing. (B) Progression of enrichment along steps of selection as evaluated by the DiVE technique [24]. The ratio between non-degraded DNA (enriched)/total DNA was calculated from image quantifications of the resulting agarose gel using the ImageJ software [41] (see material and methods). Rounds 4, 6, 8, 10 and 14 were chosen for NGS analysis. (C) Scheme depicting the workflow for sequence analysis. (D) MEME (The MEME Suite) was used to identify the most characteristic motifs in each sequenced SELEX cycle. Three motifs were the most represented in cycles 8 (motif 3), 10 (motif 2), and 14 (motif 1). Sequences from cycles 8 and 10 share a conserved region inside motifs 3 and 2, whereas this region is lost in motif 1. No representative motif was detected in cycle 4. (E) Dynamics of aptamer motifs along SELEX cycles.