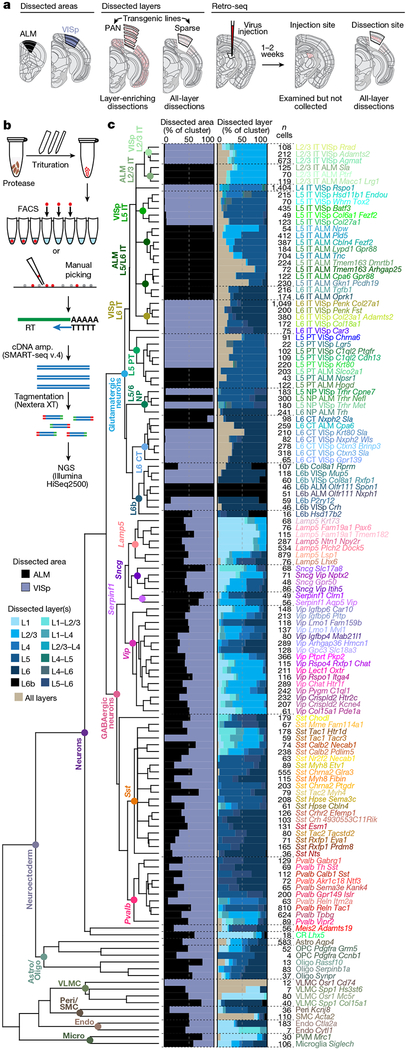
Fig. 1 |. Cell type taxonomy in ALM and VISp cortical areas.
a, Transgenically or retrogradely labelled cells and unlabelled cells were collected by layer-enriching or all-layer microdissections from the ALM or VISp. b, After dissociation, single cells were isolated by FACS or manual picking, mRNA was reverse transcribed (RT), amplified (cDNA amp.), tagmented and sequenced (next-generation sequencing, NGS). c, Clustering revealed 61 GABAergic, 56 glutamatergic, and 16 non-neuronal types organized in a taxonomy on the basis of median cluster expression for 4,020 differentially expressed genes, n = 23,822 cells and branch confidence scores > 0.4 (Extended Data Figs. 1–3). Cell classes and subclasses are labelled at branch points of the dendrogram. Bar plots represent fractions of cells dissected from the ALM and VISp, and from different layer-enriching dissections. Astro, astrocyte; CR, Cajal-Retzius cell; endo, endothelial cell; oligo, oligodendrocyte; OPC, oligodendrocyte precursor cell; peri, pericyte; PVM, perivascular macrophage; SMC, smooth muscle cell; VLMC, vascular lepotomeningeal cell; IT, intratelencephalic; PT, pyramidal tract; NP, near-projecting; CT, corticothalamic. Brain diagrams were derived from the Allen Mouse Brain Reference Atlas (version 2 (2011); downloaded from https://brain-map.org/api/index.html).