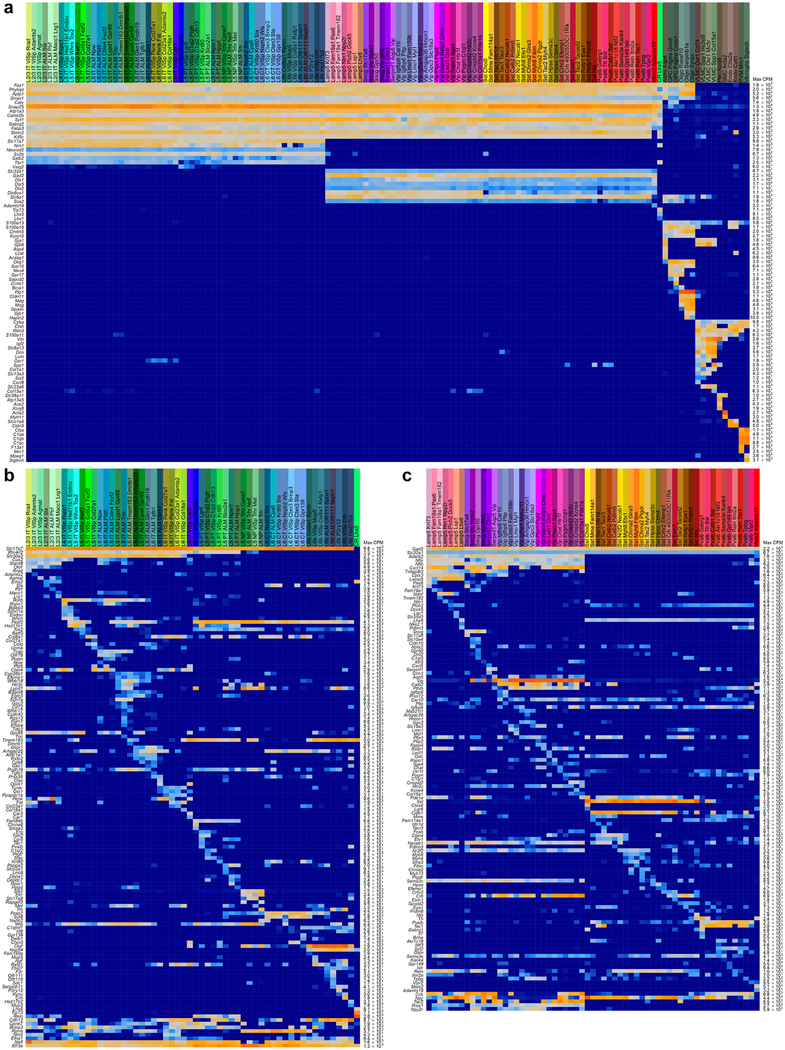
Extended Data Fig. 5 |. Markers used for cell type assignment.
a, Marker panel of 88 genes for cell classes. For each cluster, 25% trimmed mean expression values are shown (n = 23,822 cluster-assigned cells; 133 clusters). Maximum expression values (counts per million reads, CPM) are shown to the right of the heat map. Pan-neuronal markers (for example, Snap25) were used to assign neuronal type identity. Glutamatergic (for example, Slc17a7 and Slc17a6) and pan-GABAergic (for example, Gadl, Gad2 and Slc32a1) markers were used to assign glutamatergic and GABAergic identity, respectively. Known non-neuronal markers were used to assign non-neuronal identities. b, Marker panel for glutamatergic cell types. For each cluster, 25% trimmed mean expression values are shown (n = 11,905 cells; 56 clusters). Layer-specific markers were used to assign layer identity (for example, Cux2, Rorb, Deptor and Foxp2). To assign final names to types, subclass and/or layer-markers were combined with unique or other specific markers, many of which are novel. Once the identity was assigned, previously unknown genetic bases of phenotypes could be discovered. For example, for the Cajal-Retzius cell type CR-Lhx5, which has been shown by immunohistochemistry to contain glutamate but not GABA76, we show that its glutamatergic phenotype stems from the expression of mRNA encoding VGLUT2 (Slc17a6), and not from the other glutamate transporters (Slc17a7 and Slc17a8). Note that some markers that appear non-specific, provide preferential labelling of specific types when used to make random-insertion transgenic BAC lines. For example, Efr3a-cre_NO10834 specifically labels near-projecting types, although Efr3a mRNA is ubiquitously detected in all neurons. However, its expression is about fivefold higher in near-projecting types; this may contribute to preferential labelling of the near-projecting types by this BAC transgenic Cre line. c, Marker panel for GABAergic cell types. For each cluster, 25% trimmed mean expression values are shown (n = 10,534 cells; 61 clusters). GABAergic subclasses were assigned based on the expression of Lamp5, Serpinf1, Sncg, Vip, Sst and Pvalb. Final names were assigned based on unique combinations of markers.