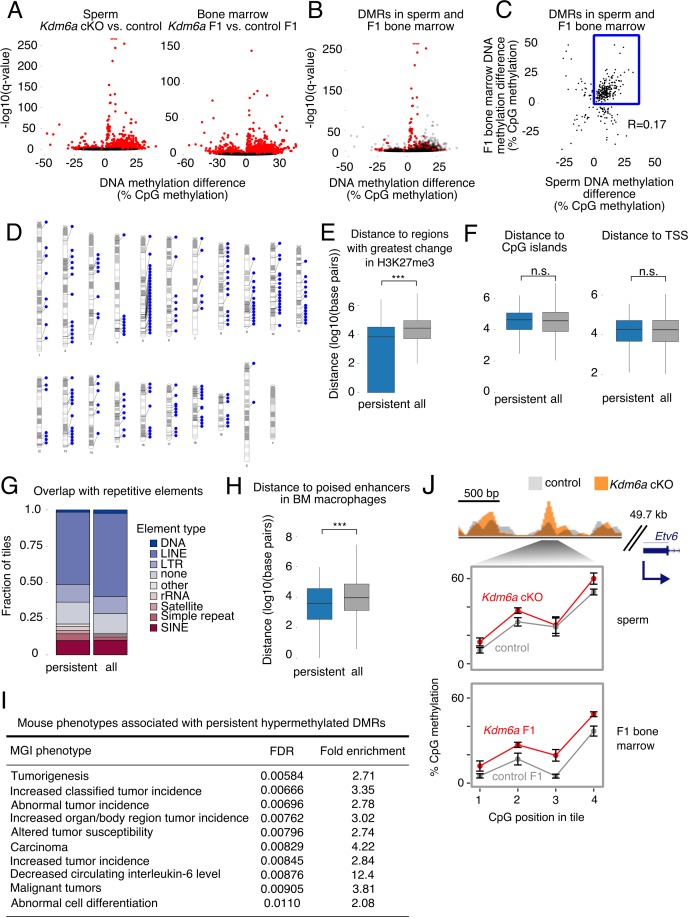
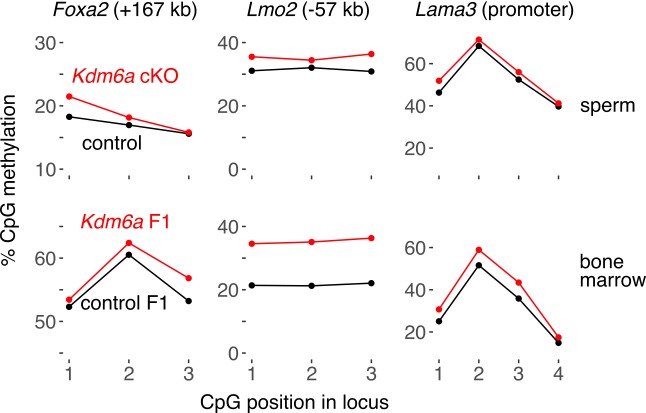
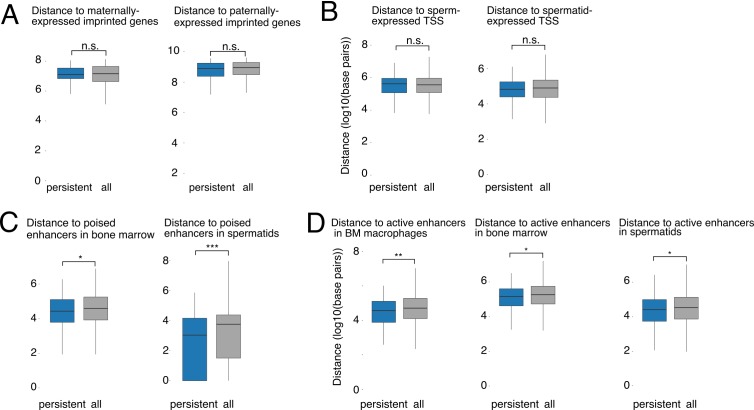
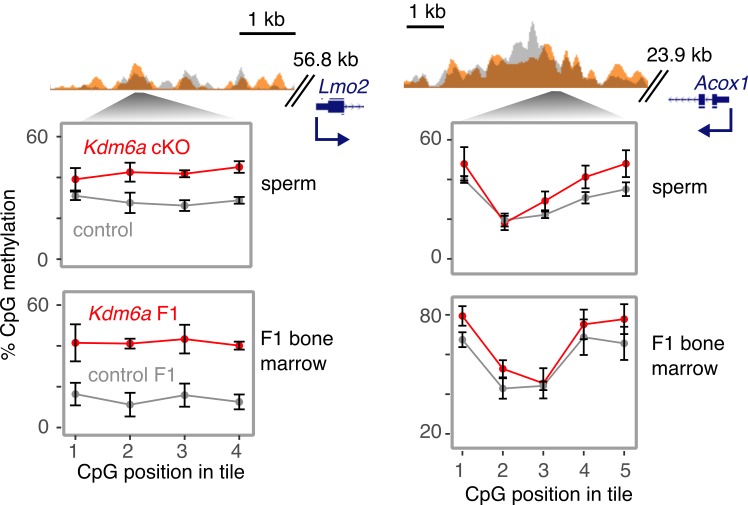
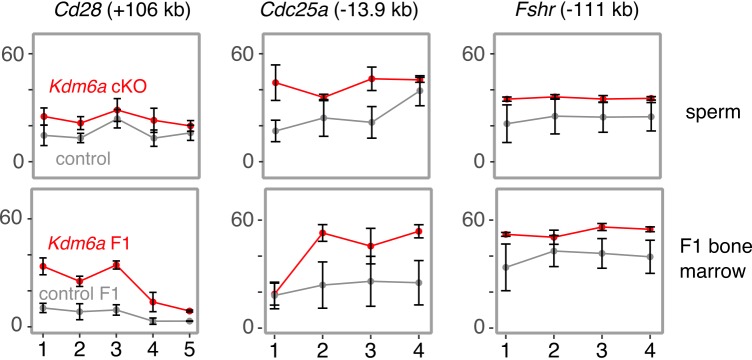
Figure 4. Persistent DMRs are associated with altered H3K27me3 and enhancer regions.
(A) Left, differentially methylated regions (DMRs) in sperm. Right, DMRs in F1 bone marrow. Red, false discovery rate (FDR) < 0.05. (B) Sperm volcano plot from (A); DMRs with FDR < 0.05 in both sperm and F1 bone marrow are in red. (C) Magnitude of DNA methylation difference (Kdm6a cKO vs. control or Kdm6a F1 vs. control F1) for the 299 DMRs shared between sperm and F1 bone marrow. Box, persistent DMRs. (D) Distribution of persistent DMRs in the mouse genome. (E) Distance from persistent DMRs to the 25% of regions with greatest change in H3K27me3 in sperm. ‘All’ refers to the complete set of tiles covered by RRBS. ***p<0.001, Mann-Whitney U test. (F) Left, distance to CpG islands. Right, distance to transcription start sites (TSS). (G) Fraction of DMRs overlapping repetitive elements. (H) Distance to poised enhancers in sorted bone marrow macrophages. ***p<0.001, Mann-Whitney U test. (I) Top 10 mouse phenotypes associated with persistent DMRs. (J) Representative persistent DMR in the enhancer of a cancer-associated gene (Etv6). Error bars, SEM of three replicates. See Figure 4—source data 1.