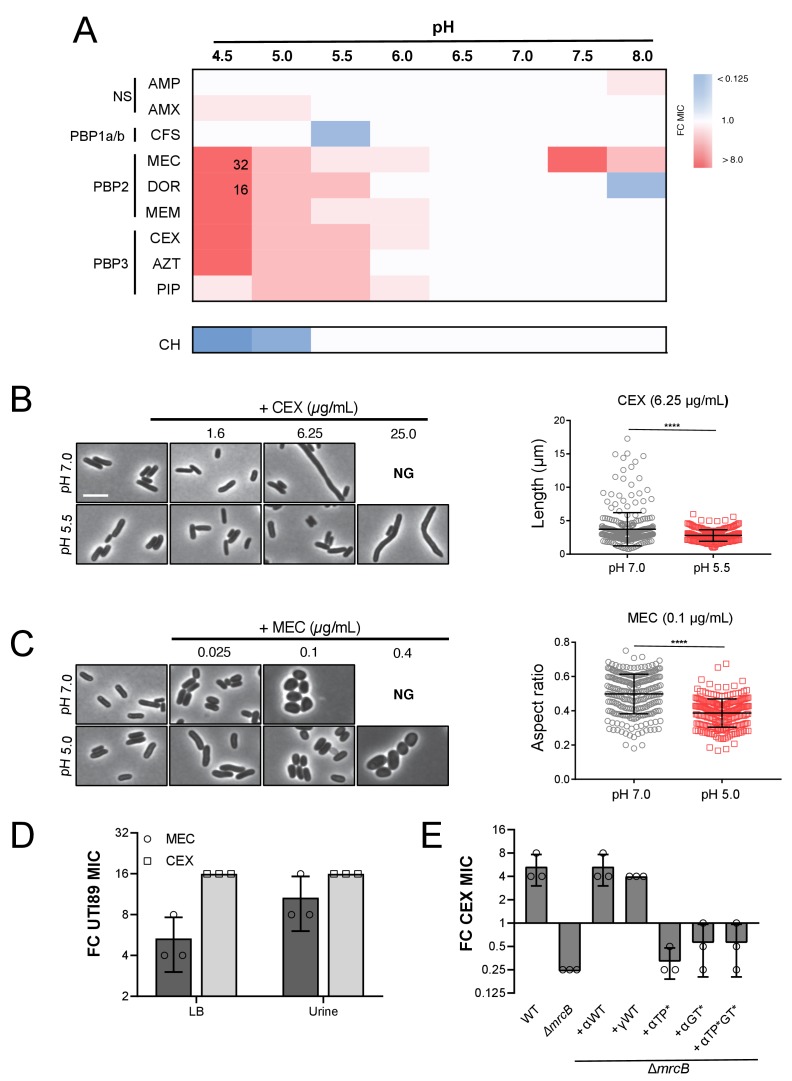
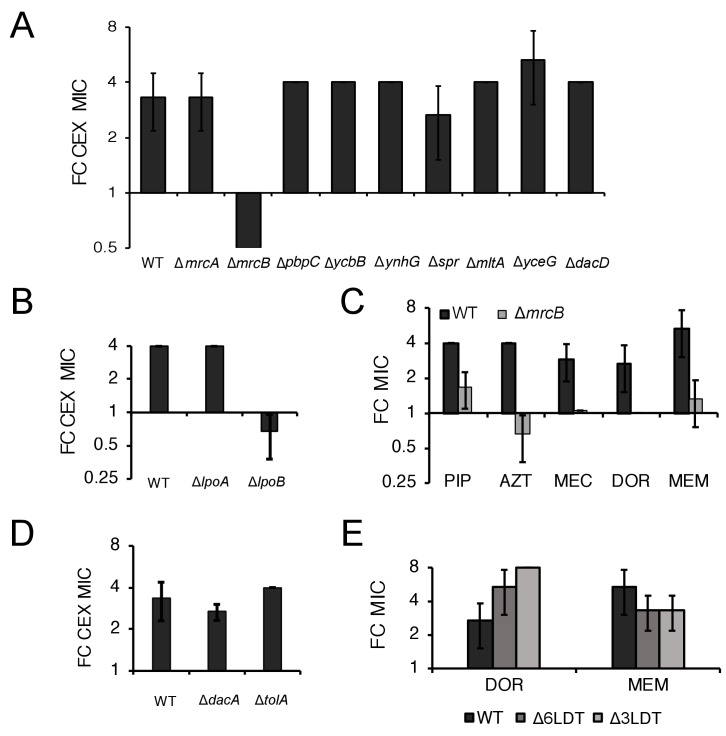
Figure 6. Intrinsic resistance to PBP2 and PBP3-targeting β-lactams at low pH.
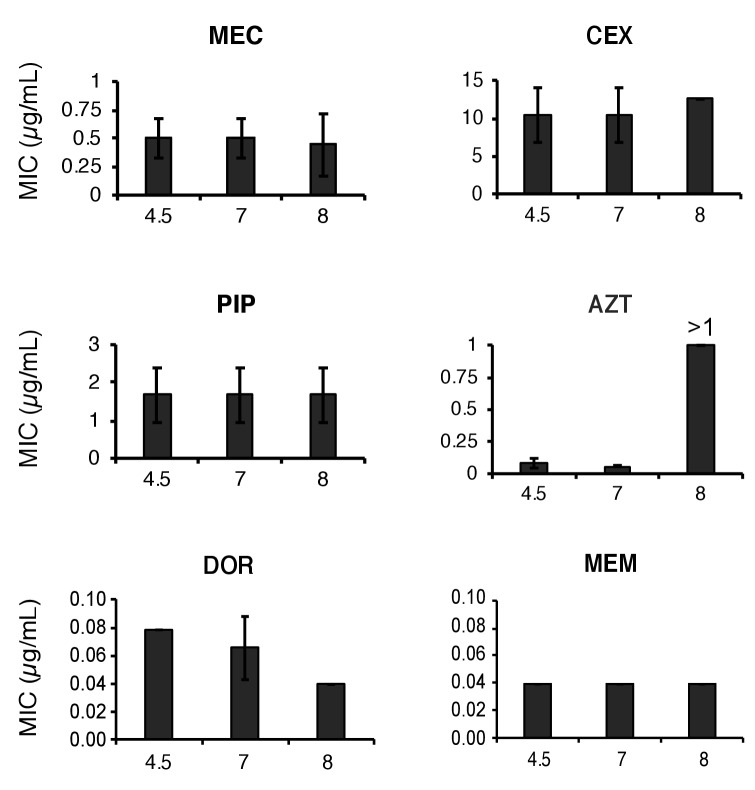
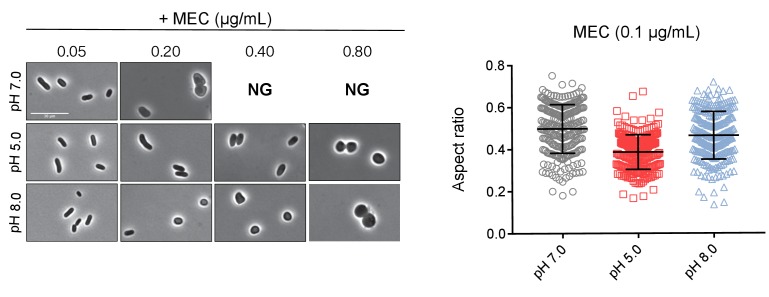
(A) Heat map summarizing fold change in minimum inhibitory concentrations (MIC) of antibiotics for strain MG1655 cultured in LB (pH 4.5–8.0) after 20 hr. Cells in heat map are colored based on median fold change (FC) in MIC at indicated pH compared to pH 7.0 from at least three biological replicates. Fold change values of >8 are indicated in black inside relevant cell. Untransformed median MIC values can be viewed in Supplementary file 4. Abbreviations for antibiotic names are as follows: AMP, ampicillin; AMX, amoxicillin; CFS, cefsulodin; MEC, mecillinam; DOR, doripenem; MEM, meropenem; CEX, cephalexin; AZT, aztreonam; PIP, piperacillin; CH, chloramphenicol. Predominant cellular PBP target is indicated to the left. (B) Representative micrographs of cells treated with PBP3 inhibitor cephalexin (CEX) at pH 7.0 and pH 5.5. Distribution of cell lengths at sub-MIC concentration at pH 7.0 and 5.5 (n = 303 and 250) are shown to the right. (B) Representative micrographs of cells treated with PBP2 inhibitor mecillinam (MEC) cultured at pH 7.0 or pH 5.0. Distribution of cell aspect ratios (width/length) at sub-MIC concentration at pH 7.0 and 5.0 (n = 250 and 250) are shown to the right. For both panels A and B, scale bar indicates 3 μm, and NG denotes ‘no growth’ observed at the indicated concentration of antibiotic. Error bars represent SD. Significance was assessed by a Kruskal-Wallis test with asterisks denoting significance as follows: ****, p<0.0001. (D) Fold change in minimum inhibitory concentration of E. coli strain UTI89 to cephalexin (CEX) and mecillinam (MEC) grown at pH 5.0 compared to pH 7.0 in broth culture and in urine. Untransformed MIC values can be viewed in Supplementary file 4. (E) Fold change in minimum inhibitory concentration to cephalexin (CEX) for indicated strains grown at pH 5.5 compared to pH 7.0. EAM696 (ΔmrcB) derivatives producing PBP1b variants were grown in the presence of 10 μM IPTG. Untransformed MIC values can be viewed in Supplementary file 5. Bars represent mean fold change in minimum inhibitory concentration ± SD across at least three biological replicates.