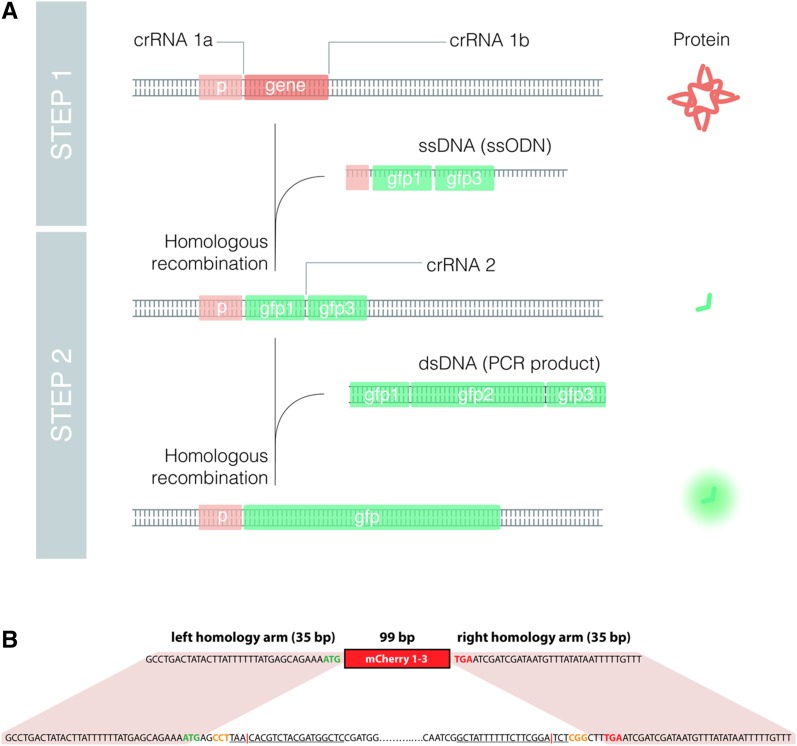
Figure 5.
Deletions and transcriptional endogenous reporters by Nested CRISPR. (A) Scheme of molecular events to generate a deletion and a transcriptional fluorescent reporter (GFP in this case) by Nested CRISPR. Two gene-specific crRNAs (crRNA 1a and crRNA 1b) and an ssODN donor are required to produce a deletion of the gene and an in-frame insertion of fragment 1–3. In the second step, RNPs contain a universal crRNA (crRNA 2) that cuts the gfp 1–3-specific targeted sequence. Then, a universal dsDNA molecule, resulting from PCR amplification of GFP, is used as a repair template to generate a transcriptional reporter. (B) Details of sequences and homology regions for simultaneous K12C11.3 knockout and mCherry 1–3 insertion. The nucleotides at the bottom represent the native sequence of the K12C11.3 gene. The entire coding sequence is removed. Nucleotides in blue represent the 5′ and 3′ untranslated regions (UTRs) while nucleotides in green and red represent the start and stop codons, respectively. Solid lines correspond to the 20-nt guide sequence, and nucleotides in orange represent PAM sequences. It can be noted that the 5′ guide sequence is in the antisense orientation. The red vertical bar represents the DSB site. The nucleotides at the top represent the sequence of the 169-bp ssODN repair template consisting of the 99-bp mCherry 1–3 sequence flanked by 35-bp homology arms.