Abstract
Linked beneficial and deleterious mutations are known to decrease the fixation probability of a favorable mutation in large asexual populations. While the hindering effect of strongly deleterious mutations on adaptive evolution has been well studied, how weakly deleterious mutations, either in isolation or with superior beneficial mutations, influence the rate of adaptation has not been fully explored. When the selection against the deleterious mutations is weak, the beneficial mutant can fix in many genetic backgrounds, besides the one it arose on. Here, taking this factor into account, I obtain an accurate analytical expression for the fixation probability of a beneficial mutant in an asexual population at mutation-selection balance. I then exploit this result along with clonal interference theory to investigate the joint effect of linked beneficial and deleterious mutations on the rate of adaptation, and identify parameter regions where it is reduced due to interference by either beneficial or deleterious or both types of mutations. I also study the evolution of mutation rates in adapting asexual populations, and find that linked beneficial mutations have a stronger influence than the deleterious mutations on mutator fixation.
Keywords: direct selection, indirect selection, mutation rates, clonal interference
ADAPTATION is driven by beneficial mutations. In a large and fully recombining population, the chance that a beneficial mutation fixes is two times its selective effect (Haldane 1927). But in an asexual population, this fixation probability is reduced either when the beneficial mutant arises on a genetic background with many deleterious mutations (background selection; Charlesworth 1994) and acquires further deleterious mutations during evolution (lineage contamination; Pénisson et al. 2017), because competing favorable mutations are present (clonal interference; Gerrish and Lenski 1998), or, in general, due to all of these reasons.
The rate of adaptation in large asexual populations has been the subject of a number of investigations, and recent experimental (Lang et al. 2013; Wiser et al. 2013; Barroso-Batista et al. 2014; Levy et al. 2015) and theoretical (Gerrish and Lenski 1998; Orr 2000; Wilke 2004; Desai and Fisher 2007; Park and Krug 2007) studies have focused on understanding the effect of competition between linked beneficial mutations on adaptation dynamics (Park et al. 2010; Sniegowski and Gerrish 2010), but these works neglect the influence of linked deleterious mutations. This is justified when mutation rates are small or selection against deleterious mutations is strong so that the beneficial mutation arises on the deleterious mutation-free genetic background (Charlesworth 2012).
However, mutation rates can rise by several orders of magnitude, at least, in adapting populations in laboratory conditions (Raynes and Sniegowski 2014), and deleterious effects can be small or moderately sized (Eyre-Walker and Keightley 2007). In these scenarios, linked deleterious mutations can substantially decrease the fixation probability of a beneficial mutation (Peck 1994; Stephan et al. 1999; Johnson and Barton 2002), and it has been shown that the fixation probability of a beneficial mutant vanishes beyond a critical mutation rate when deleterious effects are weak (Pénisson et al. 2017). In addition, superior beneficial mutations may also interfere with the spread of a beneficial mutation in sufficiently large populations (Park et al. 2010; Sniegowski and Gerrish 2010). Interference by deleterious and beneficial mutations negatively impacts the adaptation rate as has been seen, mainly numerically, in several studies (Bachtrog and Gordo 2004; Campos 2004; Campos and Wahl 2010; Jiang et al. 2011; Good and Desai 2014; Pénisson et al. 2017). However, to the best of my knowledge, accurate analytical expressions for the fixation probability and the rate of adaptation have not been obtained when deleterious effects are weak.
The above considerations may also have a bearing on the evolution of mutation rates. In a recent work, Raynes et al. (2018) have shown experimentally that asexual mutators are favored in populations larger than a critical population size, and argue that this critical value is inversely proportional to the fixation probability of a beneficial mutation. Their theoretical analysis, however, neglects any linkage effects. But as linked mutations decrease the fixation probability, it is important to ask how large the population size should be for the mutators to survive in general settings.
To address these questions, I consider a general model in which the beneficial mutant may either have the same mutation rate as that of the population in which it arises or it may be a mutator, and find an analytical expression for the fixation probability when deleterious effects are weak. I then use this result along with clonal interference theory (Gerrish and Lenski 1998) to understand the joint effect of linked deleterious and beneficial mutations on the rate of adaptation. I identify regions in the space of population size and mutation rates where either beneficial or deleterious or both kinds of mutations interfere; these results are summarized in Figure 6. I also investigate the relationship between population size and mutation rates in adapting populations, and find that clonal interference strongly affects the critical population size above which mutators are favored (see Figure 8).
Figure 6.
Regions in the space of scaled population size and scaled deleterious mutation rate where linked mutations interfere when deleterious effects are weak. The dashed and dotted lines, respectively, show (15) and (16) with beneficial mutation rate and the solid horizontal line separates the regimes where interference by deleterious mutations can be neglected and where it is effective. The boundaries separating these evolutionary regimes for are depicted, respectively, by circles, squares and triangles. The points where are not shown as the analytical expression (9) does not hold for such parameters.
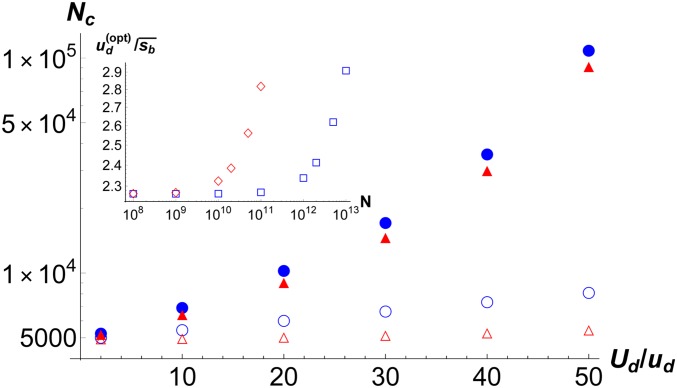
Figure 8.
Main: Critical population size above which mutators are favored when clonal interference is taken into account (filled symbols) and on neglecting it (open symbols). The data for strongly deleterious mutations (circles, ) and weakly deleterious mutations (triangles, ) are obtained by numerically solving (23) and (24), respectively. The parameters are . Inset: Scaled mutation rate at which the rate of adaptation (18) for is maximum for for the fraction of beneficial mutations (diamonds) and (square).
Models and Methods
I consider a haploid asexual population of finite size N. In every generation, an individual acquires deleterious mutations that are Poisson-distributed with mean , and each such mutation decreases its fitness by a factor . I work with large populations of size in which the Muller’s ratchet (Muller 1964; Jain 2008) operates slowly, and the population stays close to the deterministic mutation-selection equilibrium before the >first click of the ratchet.
In this resident population at mutation-selection balance, beneficial mutations that are also Poisson-distributed arise at rate . If is the total mutation rate of the population, and a fraction of all mutations are beneficial, it follows that , and for , one may write . Each beneficial mutation increases the fitness of the individual by a factor . Beneficial mutants also acquire deleterious mutations at rate that decreases their fitness by per deleterious mutation. The deleterious effects are assumed to have a fixed size , but the beneficial fitnesses are chosen from a truncated exponential distribution with mean (Eyre-Walker and Keightley 2007). Motivated by a recent work (Raynes et al. 2018), I also consider the situation in which the invading mutant is a mutator with deleterious mutation rate and beneficial mutation rate ; the fitness of a mutator individual is assumed to be the same as described above for nonmutators.
In this article, I am mainly interested in understanding how weakly deleterious mutations (as defined in the next section) affect the fixation of beneficial mutations. When the beneficial mutant is under direct selection , I analyze how linked beneficial and deleterious mutations impact the rate of adaptation in asexuals. I also study how interference by linked mutations affect the evolution of mutation rates in adapting asexual populations. Using a multitype branching process (Harris 1963; Patwa and Wahl 2008) and some ideas developed in Jain and James (2017), I first obtain an analytical expression for the fixation probability of a beneficial mutant of fixed effect size in the general model described above. I then use this result and clonal interference theory (Gerrish and Lenski 1998) to study the combined influence of deleterious and beneficial mutations on quantities of interest. Since I work in the parameter region where the deleterious effects are small, large population sizes are required to ensure that Muller’s ratchet clicks slowly. This limits the range of parameters that can be investigated numerically, but some results from individual-based simulations of the above model are reported in Supplemental Material, File S1 for , and found to agree reasonably well with the theory developed in the main text.
Results
Fixation probability of a beneficial mutant
I now proceed to calculate the fixation probability of a single beneficial mutant in a large resident population using the standard multitype branching process. As detailed in Appendix A, the fixation probability of a mutant arising in a stationary genetic background with i deleterious mutations obeys the following recursion equation,
| (1) |
In the above equation, the non-negative integer λ is the maximum number of deleterious mutations that the beneficial mutant can carry in order to have a nonzero fixation probability, and is given by
| (2) |
where denotes the maximum integer less than or equal to x. For , (1) and (2), respectively, reduce to (12) and (10) of Johnson and Barton (2002). The total fixation probability is obtained by averaging over all the genetic backgrounds,
| (3) |
where the probability that the beneficial mutation arises in a genetic sequence with i deleterious mutations is given by (Kimura and Maruyama 1966; Haigh 1978)
| (4) |
and the average number of deleterious mutations . Using the boundary condition , one can solve (1) for the fixation probability numerically (Johnson and Barton 2002).
However, it is possible to make analytical progress if one exploits the fact that, besides the fixation probability , all the mutation rates and selection coefficients are also smaller than one (Jain and James 2017). On expanding (1) in a power series about these small quantities, and retaining terms to quadratic orders (see Appendix A for details), I obtain the following equation for the fixation probability,
| (5) |
where
| (6) |
Note that the approximations employed here to arrive at (5) are similar to those used to obtain Haldane’s classical result (Haldane 1927) for fixation in a single genetic background.
Figure 1 shows the fixation probability in the genetic background with i deleterious mutations for large λ and μ, and I find that the quadratic approximation (5) somewhat overestimates the exact fixation probability (1). However, as (5) enables one to make analytical progress, I will employ the quadratic approximation for the rest of the article. Furthermore, for a given λ, the definition (6) means that . But, for weakly deleterious effects that correspond to large λ, one may write , and the recursion relation (5) as
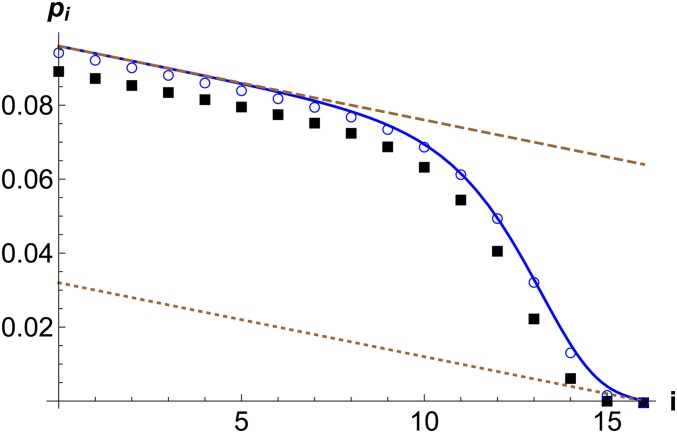
Figure 1.
Fixation probability of a beneficial mutation when it arises in a genetic background with i deleterious mutations. The points are obtained by numerically iterating the exact relation (1) (squares) and the quadratic-approximation (5) (circles). The analytical expression (8) (solid line), and the upper and lower bounds (dashed lines) given by (D.1) are also shown. The parameters are .
| (7) |
Transition in the fixation probability—a heuristic argument:
For weakly deleterious effects, although the beneficial mutant can survive in genetic backgrounds, as the background frequency is Poisson-distributed with mean and variance equal to μ [see (4)], the beneficial mutant is likely to arrive in those individuals that carry deleterious mutations lying between and . Then, if , the beneficial mutant arrives in those genetic backgrounds in which it can not survive, and hence its fixation probability is zero. On the other hand, if , the arrival of beneficial mutant occurs in those genetic backgrounds where it has a substantial chance of spreading, and the fixation probability is therefore expected to be nonzero. This simple arrival-survival argument suggests a nontrivial transition in the behavior of total fixation probability P: it is nonzero when the mutation rate and zero for . Such a transition has been observed numerically in a previous work (Johnson and Barton 2002) and shown rigorously recently (Pénisson et al. 2017); however, none of these studies provide explicit expressions for the fixation probability.
Analytical expression for fixation probability:
Equation 7 is analyzed for in Appendix B using a perturbation theory [also, see Good and Desai (2014)], and below I focus on the finite regime. In Appendix C, I show that, for large λ and μ, the fixation probability
| (8) |
The above equation has a simple interpretation: the first term on the right-hand side (RHS), which decreases linearly with increasing number of deleterious mutations, is obtained on assuming that the beneficial mutant can accumulate new deleterious mutations but does not fix on any background other than the one it arose on (see Appendix D), and is clearly a lower bound on the probability (Peck 1994; Johnson and Barton 2002; Bachtrog and Gordo 2004; Campos and Wahl 2010). The second term on the RHS of (8) that accounts for fixation on other genetic backgrounds is of a double-exponential form (i.e., ) and approaches a constant for , but decreases quickly to for . Therefore, for , the fixation probability which is an upper bound as it is obtained on assuming that the beneficial mutant does not suffer any deleterious mutations (refer Appendix D). Figure 1 shows that the upper bound is a good approximation in genetic backgrounds with few deleterious mutations, whereas the lower bound works fairly well in backgrounds with many deleterious mutations. In contrast, as Figure 1 attests, the result (8) that interpolates between these two bounds agrees well with the numerical solution of (5) for all genetic backgrounds.
As described in Appendix C, for large λ and μ, (8) yields the total fixation probability to be
| (9) |
where,
| (10) |
| (11) |
and is the error function. Figure 2 shows a comparison between the total fixation probability obtained using the quadratic approximation (5) and the analytical expression (9), and indicates that (9) overestimates the former for small and underestimates it for larger values. However, the difference is quite small, and, as discussed in Appendix C and shown in the inset of Figure 2, it decreases as the deleterious effect gets weaker. Figure 2 also shows that the upper bound on the total fixation probability given by (D.2) grossly overestimates the total fixation probability (9) for high mutation rate. As discussed earlier, the upper bound captures the effect of the deleterious mutations that the beneficial mutant inherits from the genetic background on which it arose (Charlesworth 1994), but ignores the effect of lineage contamination during evolution (Pénisson et al. 2017). The latter effect is taken care of by the last term on the RHS of (9), which is negative since the error function is an increasing function.
Figure 2.
Variation of total fixation probability P with the deleterious mutation rate of the mutator allele. The data shown in circles is obtained by numerically iterating (5) while the dashed and solid lines, respectively, show the analytical expression (9) and the upper bound (D.2). The parameters are and (main), (inset).
Equation 9 shows that in the limit , as , the total fixation probability equals for all and zero for which means that, for very weak selection against deleterious mutations, a beneficial mutant does not spread if its mutation rate exceeds its beneficial effect, but has the same chance of fixation as in a fully recombining population for any mutation rate below its beneficial fitness effect. For finite but small , the total fixation probability does not change sharply but decreases gradually from to zero when mutation rate lies between and , as argued at the beginning of this section. Thus, as supported by Figure 2, for weakly deleterious effects, the total fixation probability is well approximated by (Haldane 1927) so long as the mutation rate is sufficiently small. But with increasing mutation rate, the fixation probability decreases and eventually vanishes.
Interference effects in large asexual populations
In this section, I set to make contact with previous work, and discuss how weakly deleterious effects impact the rate of adaptation.
Fixation Probability:
Equation 9 shows that for , the probability is a function of the scaled parameters and (Johnson and Barton 2002), and decreases from one to zero with increasing . Although one expects the fixation probability to decrease with decreasing , the data in Figure 3 paints a more complex picture: when , the fixation probability decreases continuously from [see (20)] to zero with decreasing deleterious effect; this is because the beneficial mutant is likely to accumulate more deleterious mutations as the selection against deleterious mutation gets weaker. But, for , the fixation probability P is a U-shaped function: it is close to for (Haldane 1927), but, as a consequence of the transition discussed in the last section, it is given by for also. For the parameters in Figure 3, the minimum in total fixation probability occurs at . But as the result (9) is valid for large (refer Appendix C), I am unable to provide an analytical estimate for the deleterious effect that minimizes the fixation probability. For the same reason, the results in Figure 3 for the total fixation probability obtained by numerically iterating (5) and the analytical expression (9) do not agree well when λ and μ are small.
Figure 3.
Main: Variation of total fixation probability P with deleterious effect for (circles) and (triangles), and . Inset: Total fixation probability as a function of beneficial effect for . In both figures, the numerical solution of (5) is shown by points, and the analytical result (9) by lines.
The above discussion thus indicates that when the selection against deleterious mutations is weak, the loss of beneficial mutation can be mitigated if the mutation rate is small enough. To put it another way, large enough beneficial effect can overcome the deleterious effect of mutations; indeed, as the inset in Figure 3 shows, with increasing beneficial effect, the total fixation probability rises from zero toward when exceeds . As discussed below, this transition in the fixation probability plays an important role in understanding the adaptation dynamics.
Substitution rate:
In a large population of size N, if the beneficial mutations are produced at rate per individual, and the beneficial fitnesses are distributed according to the probability distribution , the expected substitution rate of beneficial mutations is given by
| (12) |
In the above equation, the effect of interfering beneficial mutations is captured by the factor , where is the average number of contending mutations with beneficial effect greater than that escape stochastic loss (Gerrish and Lenski 1998; Pénisson et al. 2017), and the distribution is a truncated exponential distribution with mean and upper bound . The latter condition is required because my results for fixation probability are derived by assuming that the selection coefficients are smaller than unity; however, it is a good approximation to replace the upper limit by infinity if (see below).
Figure 4 shows the substitution rate that is obtained numerically using (9) and (12) for two values of (and fixed μ) as a function of mean selective effect. But to get an insight into the behavior of , I consider below the limit. Although the assumption that a finite population is at mutation-selection balance breaks down when the deleterious effects are infinitesimally small (see Discussion), this limit is instructive and is a good approximation to the results for finite as Figure 4 shows.
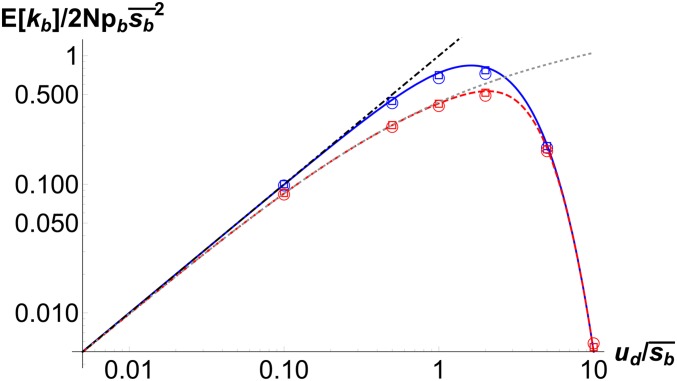
Figure 4.
Substitution rate as a function of mean beneficial effect for and . The solid and dashed lines show the substitution rate (13) for , while the points show the results obtained using (9) and (12) for (circles) and (squares). For , clonal interference occurs for , while it remains absent for all for .
The discussion in the previous section and the inset of Figure 3 suggest that, for , one may approximate the total fixation probability by a step function at so that for and zero otherwise (Johnson and Barton 2002; Pénisson et al. 2017). This results in
| (13) |
where is the expected substitution rate in the absence of any linked mutations. The integral in (13) can be numerically evaluated (see Figure 4), but on approximating it by (Wilke 2004), one arrives at
where . The condition separates the regions where interference by linked beneficial mutations can be ignored and where it matters, and gives the corresponding condition for deleterious mutations.
Equation 14a captures the effect of interference by deleterious mutations alone and shows that, for , the substitution rate decreases exponentially with increasing mutation rate, but for , there is no interference by either mutations as . This behavior is, of course, a consequence of the transition in the fixation probability, which vanishes for and equals for . Equation 14c is the substitution rate when only beneficial mutations interfere, and matches the known results in the clonal interference regime when deleterious effects are strong (Wilke 2004). The result (14b) holds when both kind of linked mutations interfere, and is obviously smaller than (14a) and (14c).
The above discussion shows that, in Figure 4, clonal interference can be ignored for the population size since for any In this case, in accordance with (14a), the substitution rate increases with and approaches . For the larger population of size , clonal interference operates when the mean beneficial effect exceeds . Then, for , only linked deleterious mutations matter, resulting in substitution rates that coincide for both values of N. But for , both deleterious and beneficial mutations affect the substitution rate and therefore it is smaller than the corresponding result for . Finally for , only clonal interference is in effect and decreases the substitution rate from .
Figure 5 shows the substitution rate for two population sizes as a function of mutation rate for a fixed fraction of beneficial mutations, and I find that it is a nonmonotonic function. For the population size for which clonal interference can be ignored, initially increases linearly with due to the increased production of beneficial mutations, and then decreases exponentially due to interference by deleterious mutations. For larger populations that satisfy , the condition yields two solutions:
| (15) |
and
| (16) |
where and are the upper and lower branch of the Lambert W function, respectively (Corless et al. 1996). This has the interesting consequence that a large population experiences clonal interference for a range of mutation rates given by , but for and , beneficial mutations fix sequentially. The data in Figure 5 for supports these conclusions as is seen to be smaller than the corresponding result for for , and is independent of N otherwise.
Figure 5.
Substitution rate as a function of deleterious mutation rate for (solid) and (dashed). The dot-dashed curve and dotted curve, respectively, show the substitution rate when there is no interference and when only clonal interference occurs. The other parameters are for (circles) and (squares). For , (15) and (16) predict that clonal interference occurs for .
In the first case , clonal interference does not click due to the low production rate of beneficial mutations. But the origin of can be traced to the behavior of the fixation probability discussed in the preceding sections: when deleterious effects are extremely weak, only beneficial effects can fix and, therefore, with increasing mutation rate, larger beneficial effects are required for adaptation to proceed. But the distribution of beneficial fitness effects is, in general, a decreasing function (Eyre-Walker and Keightley 2007) so that such large effects are rare. As a result, the large population again enters the sequential fixation regime where clonal interference is absent. This reentrant effect was demonstrated recently by Pénisson et al. (2017) when the deleterious and beneficial fitness effects are nearly equal; however, the difference between the lower and upper sequential fixation regimes was not elucidated. The above discussion and (14a)–(14c) show that mutations do not interfere in the lower regime, while only deleterious mutations hamper adaptation in the upper regime.
The boundaries separating the regions where one or both types of mutations interfere are displayed in Figure 6 for infinitesimally small deleterious effects as well as for finite , and again show that is a quite good approximation. The boundaries for finite are obtained by using the fact that clonal interference occurs when the average number of interfering mutations I with average selection coefficient (see (17) below) is above one (Pénisson et al. 2017). Note that these boundaries are independent of the mean beneficial effect in the space of scaled variables.
Adaptation rate:
I now address at what rate an asexual population adapts under the combined effect of interfering deleterious and beneficial mutations. The rate of adaptation is given by , where
| (17) |
is the average selection coefficient of fixed beneficial mutation and is smaller than one so that .
For , proceeding in a manner similar to that for the substitution rate, I find the scaled rate of adaptation to be
| (18) |
that may be approximated as (Wilke 2004)
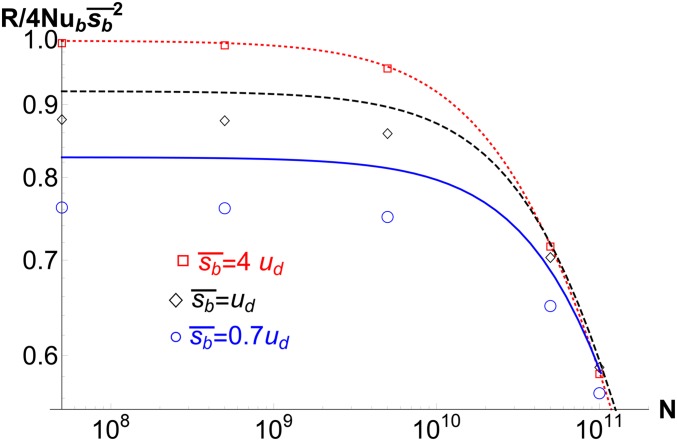
where is the adaptation rate in the absence of any interference. As discussed for the substitution rate, the behavior of the adaptation rate may also be classified into four distinct regimes. Figure 7 shows the rate of adaptation as a function of population size for various mean beneficial effects, and I find that R increases linearly with population size for and, due to clonal interference, it grows slower than N for where is the solution of . For , the adaptation rate is given by the corresponding result for lethal mutations where the effect of linked deleterious mutations can be neglected (Wilke 2004), but the interference by weakly deleterious mutations for decreases the adaptation rate further.
Figure 7.
Adaptation rate R as a function of population size N for and (points) for various . The lines show the result obtained using (18) for . The population size above which clonal interference operates is found to be for , respectively.
Optimum mutation rate:
Like substitution rate, the rate of adaptation is also a nonmonotonic function of mutation rate, and is maximum at a mutation rate . The initial increase can be attributed to the increased production of beneficial mutations and the decay to the deleterious mutational load. For small populations , from (19a), I find that the (scaled) optimum mutation rate is a solution of the equation which gives . The ratio is greater than one because the adaptation rate increases linearly but the decrease is somewhat slower than an exponential [due to the factor in the bracket on the RHS of (19a)]. For large populations , the optimum mutation rate increases, albeit mildly, with N; these results are shown in the inset of Figure 8.
Fixation of mutators in adapting asexual populations
I now study the fixation of mutators when deleterious mutations are lethal, and when they are extremely weak. In an adapting asexual population, a mutator allele can spread because it is more likely to be associated with beneficial mutations than the wild type (Sniegowski et al. 1997; Taddei et al. 1997). Since the mutator produces a beneficial mutant with probability and this beneficial mutant fixes with probability P, the mutator can hitchhike to fixation if the probability exceeds the neutral fixation probability ; this argument gives the critical population size above which the mutator can fix to be (Raynes et al. 2018).
Strongly deleterious mutations:
When the effect of deleterious mutations is large enough, the beneficial mutant can fix in only a few genetic backgrounds. In particular, only the deleterious mutation-free background matters when . Then, using (5), I immediately obtain , and
| (20) |
on using (4). When , the above equation reduces to the well known result for the fixation probability of a mutant under direct selection (Charlesworth 1994; Peck 1994). Equation 20 shows that as mutator strength (for a given ) increases, its chance of fixation decreases, since the mutant accumulates more deleterious mutations. With increasing , both the resident and the mutant carry larger number of deleterious mutations, and, therefore, the fixation probability again decreases. However, increasing the fitness advantage of the mutant enhances its chance of fixation; similarly, increasing the fitness cost of deleterious mutation means that the resident population and the mutant carry fewer deleterious mutations, and increases the fixation probability. Thus, as intuitively expected, the fixation probability increases with the selection coefficients and , but decreases with increasing mutation rates and .
Using (20) in the above argument for critical population size, I find that mutators with mutation rate cannot fix in a finite population, and
| (21) |
which shows that larger populations are required for the fixation of stronger mutators. Neglecting the reduction in fixation probability for large mutation rates, Raynes et al. (2018) predicted the critical population size which is independent of a mutator’s mutation rate. For the parameters in panel B, Figure S2 of Raynes et al. (2018), expression (21) predicts the critical population size to be 555 and 999 for mutators of strength 100 and 500, respectively, in agreement with their simulation results. For , the condition is barely satisfied, but, for mutator of strength 950, I obtain from (21), in close agreement with their simulations.
For exponentially distributed beneficial fitness effects, generalizing clonal interference theory to mutators using (20), I find that the critical population size is determined by the following equation,
| (22) |
| (23) |
where . Figure 8 shows the numerical solution of (23) for , and I find that, for strong mutators, clonal interference has a strong effect as it increases the critical population size substantially from the case where competition between beneficial mutations is neglected.
Weakly deleterious mutations:
For , as a result of the transition in the fixation probability, a population of size larger than is required for the fixation of a mutator with mutation rate . Comparing this result with (21) for lethal mutations, I find that somewhat larger populations are required for the spread of mutators when selection against deleterious mutations is strong.
The effect of clonal interference can also be included for weakly deleterious mutations, and I obtain
| (24) |
A comparison of the results for found using (23) for strongly deleterious effects and the above equation suggests that the critical population size is mildly affected by the size of the deleterious effect, see Figure 8.
Data availability
The author states that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7648805.
Discussion
In this article, I focused on understanding how weakly deleterious mutations affect the fixation of new beneficial mutations in an asexual population at mutation-selection balance. The wild-type population can stay at mutation-selection equilibrium provided the time scale over which the Muller’s ratchet clicks is large (Jain 2008). Furthermore, when a beneficial mutation arises in an individual with deleterious mutations, the subpopulation carrying the beneficial mutation is initially not in equilibrium but gets there in a time of order (Johnson 1999; Maia et al. 2003), and displaces the resident population, provided its stationary fitness exceeds that of the wild type population [see (6)], in a time of order . This argument implicitly assumes that the sweep time of the beneficial mutation is longer than the equilibration time (Johnson and Barton 2002), and requires that (on replacing i by its typical value ).
When deleterious effects are weak, as both λ and μ are large, one may ask if mutation-selection balance is possible for biologically realistic parameters. In a bacterial population of size and (Wielgoss et al. 2013), the above conditions for mutation-selection equilibrium can be met for weakly deleterious effects of size provided the beneficial effect . The above example suggests that, for biologically realistic population sizes and mutation rates, the theory presented here is applicable provided the beneficial fitness effect is larger than the deleterious one by an order of magnitude or so. Indeed, the results from stochastic simulations (shown in Figures S1 and S2) are in good agreement with the theory based on equilibrium assumption when .
Rate of adaptation in asexuals
Both deleterious (Charlesworth 2012) and beneficial (Park et al. 2010; Sniegowski and Gerrish 2010) mutations are known to interfere with the evolutionary process in asexual populations. When only deleterious mutations interfere, it is instructive to compare the extreme cases in which selection against deleterious mutations is very strong or very weak. In the former case, a beneficial mutation with effect fixes with probability (20) that decreases with increasing mutation rate from the classical result , and approaches zero asymptotically. But, in the latter case, while the corresponding fixation probability also decreases with , the catastrophic reduction to zero occurs at a finite mutation rate that is equal to (see Figure 2). This transition in the fixation probability may be understood by the following back-of-the-envelope calculation: since the relative fitness of a beneficial mutant with i deleterious mutations, each of deleterious effect , is , the mutant fixes with the probability (Haldane 1927). But the average number of deleterious mutations in the population is while the mutant can tolerate at most deleterious mutations to avoid extinction. Thus, when , the fixation probability is (on replacing i by its average μ) and zero otherwise as has been shown in previous work (Johnson and Barton 2002; Pénisson et al. 2017). Although this simple argument captures the qualitative behavior of the fixation probability, a more refined calculation that agrees quantitatively with numerical (Figure 1, Figure 2, and Figure 3) and simulation (Figures S1 and S2) results yields the expression (9) for the fixation probability.
The above transition plays a key role in understanding the adaptation dynamics when both deleterious and beneficial mutations interfere with the fixation of a beneficial mutation. As summarized in Figure 6, when the scaled mutation rate (with being the mean beneficial effect), interference by deleterious mutations can be ignored and one arrives at familiar results for the adaptation rate (Orr 2000; Wilke 2004; Sniegowski and Gerrish 2010), viz., it increases linearly with population size for small populations and logarithmically for larger ones [see (19a) and (19c)]. However, when the scaled mutation rate is above one, linked deleterious mutations begin to interfere. Then for large mutation rates, the substitution rate becomes essentially independent of the population size, unlike in the case where interference by deleterious mutations is neglected [cf. Figure 5 with Figure 3 of Wilke (2004)]. Because of the transition in the fixation probability discussed above, at high mutation rates, only large beneficial effects stand a chance to spread, but such effects are rare and therefore competing beneficial mutations become unavailable and clonal interference disappears (Pénisson et al. 2017).
Evolution of mutation rates in large asexual populations
Although mutations drive the evolutionary process by creating novel genetic variation in a population, high mutation rates are generally not favored as mutators produce, on average, more deleterious mutations. Thus, in a well adapted population where beneficial mutations are not available, antimutators that lower the mutation rate are likely to be favored, and the mutation rate will keep decreasing until the indirect selective advantage conferred due to reduced deleterious load becomes comparable to the strength of the genetic drift, (Lynch 2010; Lynch et al. 2016). Thus, in an adapted population, mutation rate and population size are negatively correlated, and the quantitative relationship between them has been worked out for both strongly and weakly deleterious mutations (James and Jain 2016; Jain and James 2017).
On the other hand, in an adapting population, besides deleterious mutations, a mutator produces more beneficial mutations, and, in an asexual population, it may hitchhike to fixation with them thus increasing the mutation rate (Sniegowski et al. 1997; Taddei et al. 1997). The (indirect) advantage offered by a mutator due to increased beneficial production rate, however, decreases with increasing mutator strength, and, therefore, a strong mutator may overcome the loss due to random genetic drift in sufficiently large populations (Raynes et al. 2018). Thus in an adapting population, mutation rate and population size are positively correlated.
From the analysis here based on this argument, I arrive at the following main conclusions: First, I find that the size of the deleterious effect does not significantly affect the critical population size needed for the fixation of mutators in adapting populations. In contrast, in the absence of beneficial mutations, the size of deleterious effect changes the quantitative relationship between population size and mutation rate [see (14) of James and Jain (2016)]. Second, for both strongly and weakly deleterious effects, clonal interference increases the critical population size above which mutators are favored quite substantially (see Figure 8). This inflation in critical population size occurs because, as mentioned above, larger populations are needed for the fixation of stronger mutators. But beneficial mutations compete in large populations thus decreasing the fixation probability of a beneficial mutation-carrying mutator, and therefore even larger populations are required for the mutator to escape stochastic loss.
A similar effect is also seen for the optimal mutation rate at which the rate of adaptation is maximum when the population enters the clonal interference regime. As the inset of Figure 8 shows, the optimum mutation rate increases mildly with N; this is in contrast to the case when deleterious mutations are lethal where the optimum mutation rate remains independent of population size (Orr 2000). I also note that when interference by deleterious mutations can be ignored, the population size above which clonal interference comes into play decreases with increasing mutation rate, but it increases with mutation rate when deleterious mutations interfere, see Figure 6.
Limitations and open questions
In this article, I considered an asexual population at mutation-selection balance. But the equilibrium assumption does not hold, for example, following a selective sweep or when Muller’s ratchet clicks rapidly (Johnson and Barton 2002; Bachtrog and Gordo 2004; Campos 2004); a detailed analysis of adaptation rate in such nonequilibrium populations is beyond the scope of this article and will be addressed in a future work. I worked in the parameter regime where the production rate of beneficial mutations is small thereby neglecting the possibility of multiple-mutations in the same individual (Desai and Fisher 2007; Park and Krug 2007). Extending the present work to include the combined effect of clonal interference and such multiple-mutations (Good et al. 2012), besides the interference by weakly deleterious effects, would be interesting.
Here, I focused on the spread of beneficial mutations in a fully linked genome, and an important direction for the future will be to explore the effect of recombination on the results obtained in this work (Weissman and Barton 2012). A beneficial mutation cannot fix beyond a critical mutation rate in an asexual population (Johnson and Barton 2002; Pénisson et al. 2017). But, as the linked alleles dissociate over time in a recombining population, it would be interesting to find how the fixation probability increases, particularly in the vicinity of the critical mutation rate, with increasing recombination rate. More generally, for sufficiently high recombination rate, interference from linked mutations are expected to disappear or get considerably weakened. But to what extent small recombination rates decrease the hindering effects of linked beneficial and deleterious mutations remains a question for the future.
Acknowledgments
I thank Mark Kirkpatrick for helpful discussions during the early stages of this work. I am also grateful to Brian Charlesworth, Joachim Krug, Lindi Wahl, and three anonymous reviewers for useful comments on an earlier version of the manuscript. I acknowledge funding from the Department of Biotechnology-Jawaharlal Nehru Centre for Advanced Scientific Research (DBT-JNCASR) grant “Life science research, education and training at JNCASR” (BT/INF/22/SP27679/2018), Government of India.
Appendix
Appendix A Quadratic Approximation for Fixation Probability
Let denote the probability that a beneficial mutant arising at time t in a wild type individual with i deleterious mutations eventually gets fixed. If the mutant gives rise to n offspring in the next generation with probability and j deleterious mutations occur in an offspring with probability , the extinction probability where is the probability that all n lineages go extinct (Johnson and Barton 2002). Here, the mutation probability , and the offspring number is also Poisson-distributed with mean equal to the fitness of the mutant relative to the average fitness of the resident population, so that . For a slowly clicking ratchet, as the resident population remains at mutation-selection balance, the beneficial mutation arises and fixes in a stationary background, and, therefore, the fixation probability is independent of time. Putting these pieces together results in the recursion relation (1) for the fixation probability. Furthermore, in order to have a nonzero fixation probability , the fitness of the displacing population with at least i deleterious mutations must exceed that of the resident population, i.e., which means that , where λ is given by (2).For , it is a good approximation to assume that an offspring suffers at most one deleterious mutation, so that one may write . For the same reason, one may also ignore the contribution from those lineages in which more than one offspring undergoes mutation, and write
| (A.1) |
| (A.2) |
where I have used that the fixation probability is small because the relative fitness of the mutant is close to one when . Using these approximations in (1) and (2) leads to (5) and (6), respectively, in the main text.
Appendix B Fixation Probability for
When but , the ratios . I therefore expand in a power series in the small parameter and write . Substituting this in (7), and keeping terms to linear order in , one obtains
| (B.1) |
| (B.2) |
where the last expression follows on using that for , the mean of the frequency distribution is smaller than λ, and, therefore, fitness classes that contribute to the sum (3) for total fixation probability are smaller than λ . If now , one may write the total fixation probability as with and arrive at
| (B.3) |
For , one can approximate the frequency distribution by a Gaussian distribution with mean and variance μ. On carrying out the integral , I again obtain (B.3).
Appendix C Fixation Probability for Finite
To find the fixation probability of a beneficial mutant for and finite , I essentially follow the treatment in Jain and James (2017). I first note that when the background mutation , the second term on the left-hand side (LHS) of (7) may be ignored. Then the solution that obeys is simply given by
| (C.1) |
where I have used . I now write in (7) to obtain
| (C.2) |
| (C.3) |
where the last expression follows on using that . If one now sets the LHS of (C.3) to zero, one gets which is a good approximation for small or large μ, as can be seen by using the approximate solution on the RHS of (C.3). Adding the above expressions for and yields (8).To find the total fixation probability, as before, I approximate by a Gaussian distribution with mean and variance equal to μ. The integral over is exactly doable, and gives the first two terms on the RHS of (9). To handle the term containing , I approximate it by where is the Heaviside theta function and which yields
| (C.4) |
and the integral over the Gaussian distribution leads to the last term on the RHS of (9).
Appendix D Bounds on Fixation Probability
Since the probability , the first term on the RHS of (7) lies between zero and . This implies that
| (D.1) |
These bounds have a simple meaning: the lower bound is obtained on assuming that the beneficial mutant can mutate but does not fix on any background other than the one it arose on , while on setting the deleterious mutation rate of the mutant to zero , the upper bound given by twice the relative fitness of the mutant with respect to the average fitness of the resident population (Haldane 1927) is obtained. These bounds can also be seen from the full recursion relation (1) as follows: if one neglects and allows only the unmutated fraction of the offspring population to contribute to fixation, the lower bound is obtained (Peck 1994; Johnson and Barton 2002). The upper bound is, perhaps, a bit subtle to see when ; here, one is assuming that the offspring of the beneficial mutant do not suffer a deleterious mutation with probability equal to one, which is implemented by setting the mutation rate to be zero under the summation over the genetic backgrounds in (1), but not in the fitness of the wild-type population.Although exact expressions for the bounds on total fixation probability can be found by carrying out the sum in (3), they are not particularly illuminating. However, for large μ, the Poisson distribution can be approximated by a Gaussian with mean and variance μ, and the sum in (3) by an integral. On performing the integrals, one finds that for , the bounds on the fixation probability are given by
| (D.2) |
where and is the error function, which has the property that .Then, it is easy to see that, in the limit , there is a transition in the behavior of the above bounds at . Specifically, the lower bound on the total fixation probability decreases linearly with as for and is zero for . The upper bound, on the other hand, equals for all but vanishes for .
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7648805.
Communicating editor: L. Wahl
Literature Cited
- Bachtrog D., Gordo I., 2004. Adaptive evolution of asexual populations under Muller’s ratchet. Evolution 58: 1403–1413. 10.1111/j.0014-3820.2004.tb01722.x [DOI] [PubMed] [Google Scholar]
- Barroso-Batista B., Sousa A., Lourenco M., Bergman M. L., Sobral D., et al. , 2014. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 10: e1004182 10.1371/journal.pgen.1004182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos P. R., 2004. Fixation of beneficial mutations in the presence of epistatic interactions. Bull. Math. Biol. 66: 473–486. 10.1016/j.bulm.2003.08.012 [DOI] [PubMed] [Google Scholar]
- Campos P. R., Wahl L. M., 2010. The adaptation rate of asexuals: deleterious mutations, clonal interference and population bottlenecks. Evolution 64: 1973–1983. 10.1111/j.1558-5646.2010.00981.x [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63: 213–227. 10.1017/S0016672300032365 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22. 10.1534/genetics.111.134288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless R., Gonnet G., Hare D., Jeffrey D., Knuth D., 1996. On the Lambert W function. Adv. Comput. Math. 5: 329–359. 10.1007/BF02124750 [DOI] [Google Scholar]
- Desai M., Fisher D., 2007. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798. 10.1534/genetics.106.067678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103: 127–144. 10.1023/A:1017067816551 [DOI] [PubMed] [Google Scholar]
- Good B., Desai M., 2014. Deleterious passengers in adapting populations. Genetics 198: 1183–1208. 10.1534/genetics.114.170233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good B. H., Rouzine I. M., Balick D. J., Hallatschek O., Desai M., 2012. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl. Acad. Sci. USA 109: 4950–4955. 10.1073/pnas.1119910109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J., 1978. The accumulation of deleterious genes in a population - Muller’s ratchet. Theor. Popul. Biol. 14: 251–267. 10.1016/0040-5809(78)90027-8 [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. Part V: selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. 10.1017/S0305004100015644 [DOI] [Google Scholar]
- Harris T., 1963. The Theory of Branching Processes. Springer-Verlag, Berlin; Heidelberg: 10.1007/978-3-642-51866-9 [DOI] [Google Scholar]
- Jain K., 2008. Loss of least-loaded class in asexual populations due to drift and epistasis. Genetics 179: 2125–2134. 10.1534/genetics.108.089136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K., James A., 2017. Fixation probability of a nonmutator in a large population of asexual mutators. J. Theor. Biol. 433: 85–93. 10.1016/j.jtbi.2017.08.027 [DOI] [PubMed] [Google Scholar]
- James A., Jain K., 2016. Fixation probability of rare nonmutator and evolution of mutation rates. Ecol. Evol. 6: 755–764. 10.1002/ece3.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Xu Z., Li J., Shi Y., Wu W., et al. , 2011. The influence of deleterious mutations on adaptation in asexual populations. PLoS One 6: e27757 10.1371/journal.pone.0027757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., 1999. The approach to mutation-selection balance in an infinite asexual population, and the evolution of mutation rates. Proc. Biol. Sci. 266: 2389–2397. 10.1098/rspb.1999.0936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., Barton N., 2002. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Maruyama T., 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54: 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Rice D. P., Hickman M. J., Sodergren E., Weinstock G. M., et al. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574. 10.1038/nature12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. F., Blundell J. R., Venkataram S., Petrov D. A., Fisher D. S., et al. , 2015. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519: 181–186. 10.1038/nature14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2010. Evolution of the mutation rate. Trends Genet. 26: 345–352. 10.1016/j.tig.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Ackerman M. S., Gout J.-F., Long H., Sung W., et al. , 2016. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 17: 704–714. 10.1038/nrg.2016.104 [DOI] [PubMed] [Google Scholar]
- Maia L. P., Botelho D. F., Fontanari J. F., 2003. Analytical solution of the evolution dynamics on a multiplicative-fitness landscape. J. Math. Biol. 47: 453–456. 10.1007/s00285-003-0208-8 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1964. The relation of recombination to mutational advance. Mutat. Res. 1: 2–9. 10.1016/0027-5107(64)90047-8 [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2000. The rate of adaptation in asexuals. Genetics 155: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Krug J., 2007. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA 104: 18135–18140. 10.1073/pnas.0705778104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Simon D., Krug J., 2010. The speed of evolution in large asexual populations. J. Stat. Phys. 138: 381–410. 10.1007/s10955-009-9915-x [DOI] [Google Scholar]
- Patwa Z., Wahl L., 2008. The fixation probability of beneficial mutations. J. R. Soc. Interface 5: 1279–1289. 10.1098/rsif.2008.0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck J., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pénisson S., Singh T., Sniegowski P., Gerrish P., 2017. Dynamics and fate of beneficial mutations under lineage contamination by linked deleterious mutations. Genetics 205: 1305–1318. 10.1534/genetics.116.194597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynes Y., Sniegowski P., 2014. Experimental evolution and the dynamics of genomic mutation rate modifiers. Heredity 113: 375–380. 10.1038/hdy.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynes Y., Wylie C. S., Sniegowski P. D., Weinreich D. M., 2018. Sign of selection on mutation rate modifiers depends on population size. Proc. Natl. Acad. Sci. USA 115: 3422–3427. 10.1073/pnas.1715996115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. D., Gerrish P. J., 2010. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1255–1263. 10.1098/rstb.2009.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. D., Gerrish P. J., Lenski R., 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387: 703–705. 10.1038/42701 [DOI] [PubMed] [Google Scholar]
- Stephan W., Charlesworth B., McVean G., 1999. The effect of background selection at a single locus on weakly selected, partially linked variants. Genet. Res. 73: 133–146. 10.1017/S0016672399003705 [DOI] [Google Scholar]
- Taddei T., Radman M., Maynard-Smith J., Toupance B., Gouyon P. H., et al. , 1997. Role of mutator alleles in adaptive evolution. Nature 387: 700–702. 10.1038/42696 [DOI] [PubMed] [Google Scholar]
- Weissman D. B., Barton N. H., 2012. Limits to the rate of adaptive substitution in sexual populations. PLoS Genet. 8: e1002740 10.1371/journal.pgen.1002740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgoss S., Barrick J. E., Tenaillon O., Wiser M. J., Dittmar W. J., et al. , 2013. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. USA 110: 222–227. 10.1073/pnas.1219574110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke C. O., 2004. The speed of adaptation in large asexual populations. Genetics 167: 2045–2053. 10.1534/genetics.104.027136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser M. J., Ribeck N., Lenski R. E., 2013. Long-term dynamics of adaptation in asexual populations. Science 342: 1364–1367. 10.1126/science.1243357 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The author states that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7648805.