Abstract
DNA methylation and histone modifications such as methylation, acetylation, and phosphorylation, are two types of epigenetic modifications that alter gene expression. These additions to DNA regulatory elements or to the tails of histones can be inherited or can also occur de novo. Since epigenetic modifications can have significant effects on various processes at both the cellular and organismal level, there has been a rapid increase in research on this topic throughout all fields of biology in recent years. However, epigenetic research is relativity new for the inner ear field, likely due to the limited number of cells present and their quiescent nature. Here, we provide an overview of methods used to detect DNA methylation and histone modifications with a focus on those that have been validated for use with limited cell numbers and a discussion of the strengths and limitations for each. We also provide examples for how these methods have been used to investigate the epigenetic landscape in the inner ear and related tissues.
Keywords: DNA methylation, histone methylation, histone acetylation, histone phosphorylation
Introduction
The term epigenetics refers to changes in gene expression which can be mitotically or meiotically heritable, but do not arise from changes to actual DNA sequences. Example mechanisms of epigenetic regulation include DNA modifications, chromatin structural dynamics, histone modifications, microRNAs, and non-coding RNAs. In addition to being heritable, epigenetic changes can occur de novo in response to environmental factors, intra- and extra-cellular signaling, normal cellular processes such as development or aging, and pathological conditions. Epigenetic changes can have significant effects on numerous cellular and organismal processes including, but not limited to: transcription, replication, recombination, cell cycle entry and progression, stemness/multipotency, differentiation, damage repair, metabolism, survival, migration, morphology/polarity, and physiology (Chao and D’Amore, 2008; Feinberg, 2007; Greer and Shi, 2012). As such, there has been a dramatic increase in research focused on epigenetic modifications and epigenetic regulation of gene expression in all fields of biology.
The present review is focused on the most common DNA modification, methylation, and the best characterized histone modifications: acetylation, methylation, and phosphorylation. For DNA methylation, methyl groups are added by DNA methyltransferases (DNMTs), most commonly at cytosine residues that are followed by a guanine residue (or CpG dinucleotides). This results in decreased gene expression by either recruiting proteins that condense the chromatin or by preventing transcription factors from binding (Bird, 2002; Bird and Wolffe, 1999). DNA methylation is relatively stable compared to histone modifications and therefore more likely to be heritable. However DNA demethylation can occur passively by successive rounds of cell division in the absence of DNMTs, or actively by ten-eleven translocation (TET) enzymes (Kohli and Zhang, 2013).
Histone proteins, which form complexes that wind and unwind DNA into condensed or open states, can accept post-translational modifications on their N-terminal tails, resulting in either increased or decreased gene expression depending on the modification, its location, and the cellular context. While the majority of histone modification studies have focused on histone proteins H3 and H4, histone proteins H1, H2A, and H2B, as well as several of the known histone variants (e.g. H2AX), can also be modified. Among the better studied modifications are histone acetylation, methylation, and phosphorylation. Histone acetylation occurs when histone acetyltransferases (HATs) catalyze the transfer of an acetyl group from acetyl coenzyme A to conserved lysine residues. This removes lysine’s positive charge, decreasing the affinity between DNA and histones and opening the chromatin. In general, this allows transcription factors and polymerases easier access to promoter regions to enhance gene expression. Histone deacetylates (HDACs) remove acetyl groups, which promotes chromatin condensation and generally decreases transcription (Eberharter and Becker, 2002).
Methylation of histones can occur at basic amino acid residues (lysine, arginine, and histidine). However, methylation of histidine appears to be rare and the methylation of arginine and lysine residues are therefore more commonly studied (Greer and Shi, 2012). To date, several methyltransferases have been characterized for their ability to methylate various residues on histone tails. These generally belong to the families of SET-domain containing proteins, DOT1-like proteins, and the protein arginine N-methyltransferases (PRMTs). The first two methylate lysine residues while the latter methylates arginines. Demethylation is also accomplished by a number of enzymes, some known and some unknown. While arginine demethylases are not well elucidated, a number of lysine demethylases that act on histone tails have been characterized. The majority of these belong to two families: the amine oxidases and the jumonji C (JmjC)-domain-containing dioxygenases (Biswas and Rao, 2018; Greer and Shi, 2012). It is important to note that many of these enzymes are not specific to histones, but can methylate or demethylate other proteins as well (Carlson and Gozani, 2016) Thus, experiments that knockout or inhibit the activity of specific enzymes that alter the methylation status of histones may reveal pleiotropic effects.
Unlike histone acetylation, which has the general feature of accessible chromatin and increased transcriptional activity, histone methylation is more complex. The site of methylation, as well as the number of methyl groups added at an individual residue, and whether the methyl groups are symmetric or asymmetric, can have opposing effects on chromatin condensation and transcription. Perhaps the three most well-studied histone modifications are methylation of lysines 4, 9, and 27 on histone protein H3 (H3K4, H3K9, and H3K27, respectively). Trimethylation of H3K4 (H3K4me3) is generally associated with increased transcriptional activity, while H3K9me2, H3K9me3, and H3K27me3 are associated with transcriptional repression. While the simple example of these three methylation sites illustrates how the outcome is dependent upon which residue is methylated, it is also known that the number of methyl groups on an individual residue can have opposing effects on transcription. For example, while H3K9me2 and H3K9me3 are generally repressive, H3K9 monomethylation (H3K9me) is generally associated with open chromatin and active transcription (Barski et al., 2007). In addition, even some modifications thought to be acting in one way, can, in certain cases, act in the opposite manner. H3K4me3, which is generally associated with transcriptional activation, has been shown to repress transcription under certain circumstances (Shi et al., 2006). While the effects of histone methylations are admittedly complex, this complexity suggests that histone modifications may work in a coordinated fashion. The combination of methylations, acetylations, and/or other modifications at multiple sites, rather than just the methylation status of one specific residue, may be what determines the overall effect on chromatin structure and transcriptional activity (Bernstein et al., 2006).
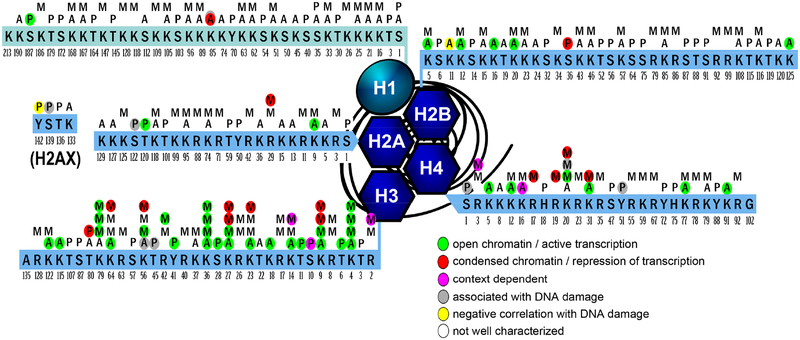
Although acetylation and methylation are the most heavily studied histone modifications, there are a number of residues on histone proteins that can be phosphorylated, including serines, threonines, tyrosines, and histidines. While the list of kinases and phosphatases shown to phosphorylate or dephosphorylate histone residues is too long to describe here (see Rossetto et al., 2012), there are also many writer and eraser enzymes that have yet to be identified. Also, similar to what is seen with the enzymes that methylate and demethylate histone residues, the enzymes that affect histone phosphorylation are generally promiscuous and exert enzymatic activity on other proteins as well (Baek, 2011). While less is known about the effects of phosphorylation at many histone sites, certain phosphorylations have been associated with proliferation or cell death. Figure 1 summarizes the residues on histone proteins H1, H2A, H2AX,H2B, H3, and H4 that are known to be acetylated, methylated, or phosphorylated and, if known, the general effects of these modifications.
Figure 1:
Post-translational modifications of histones. Sites known to undergo acetylation (A), methylation (M), and/or phosphorylation (P) are shown for the four canonical histone core proteins (H2A, H2B, H3, and H4, blue hexagons) as well as for the linker histone H1 (teal oval) and the histone variant H2AX (which extends beyond the 129 amino acids of the canonical H2A subunit). Modifications for which the function has been described are color coded: green labels marks most often linked to chromatin openness or active transcription and red labels marks most often associated with chromatin condensation and transcriptional silencing. While it is possible that all of the modifications (including those shown as red or green) may act in a context dependent manner, those labeled with magenta have roughly equal numbers of reports suggesting both actions. Modifications in gray have been described as primarily occurring in close proximity to double strand breaks or other DNA damage, while those coded yellow are markedly absent when there is DNA damage in close proximity. For those modifications that are not assigned a color, relatively little is known about their function. It is important to note that many more residues that could potentially be modified exist on these histone proteins, but only those where an acetylation, methylation, or phosphorylation have been experimentally demonstrated are shown, except for the N’ terminal amino acids which have been included on each tail to indicate their overall length. Additionally, residues known to be modified in other ways (e.g. ubiquitination or ribosylation) have been excluded, as have histone variants other than H2AX. Methylation states are represented such that a single methylation mark (“M”) denotes a residue that has been shown to be methylated, but differences between mono-, di-, or tri-methylation have not been described. For those residues with 2 or 3 “M” marks, monomethylation is represented by the bottom “M” closest to the peptide sequence, trimethylation is in the uppermost position, and dimethylation is in the middle. Finally, it is also important to note that color coding of functions is based on either currently accepted general function, or, in some cases, on limited publications that exist for a particular modification. Therefore, one cannot assume the represented function will occur in all cases, especially for modifications that have recently been characterized. As an example of this, Shi et al., 2006 demonstrated a silencing function for H3K4me3, however, there are many more reports which show H3K4me3 as a mark of active transcription which is its more generally accepted function, so it is shown as green in the figure.
Despite the rapid pace of advancement in epigenetic studies across biology, epigenetic research in the inner ear is relatively new. The few epigenetic studies that have been conducted using cells from the inner ear or related tissues have shown that DNA methylation and histone modifications play critical roles in development, aging, hair cell (HC) survival, response to insult, and HC regeneration (reviewed in Doetzlhofer and Avraham, 2017). However based on studies from other organs and cell types, it is likely that changes to the epigenetic landscape will affect nearly every cellular process and every cell type within the inner ear. Understanding these epigenetic changes will be critical to developing therapies to treat or prevent hearing loss. Yet, researchers studying the inner ear and associated tissues must face the added challenges of tissue inaccessibility, small cell populations, and heterogeneity of cell types. These can be significant obstacles as most of the techniques available for epigenetic research were developed using cell lines and are routinely carried out with millions, if not billions, of homogeneous cells in each sample. In stark contrast to this, the mouse cochlea, a common model for the study of audition, is encased in bone, contains less than 4,000 HCs on average (Ehret and Frankenreiter, 1977), and exhibits a high rate of cell death upon dissociation. Furthermore, mechanosensory organs like the organ of Corti are comprised of several different cell types with differing morphologies, functions, and transcriptomes. It is likely, therefore, that each cell type possesses unique epigenomic features as well. Thus, any attempts to investigate epigenetic modifications within or between subsets of cochlear cell types (e.g. HC vs. supporting cell (SC), or apical vs. basal turns of the cochlea) not only require cell sorting, but result in a further reduction in the number of viable cells that can be obtained for study. Furthermore, cells in the mammalian inner ear are largely quiescent which has required much additional effort to generate cell culture models (Chang et al., 2015; Koehler and Hashino, 2014; Walters et al., 2016). While some cell culture models have been developed, it is important to consider that they are still largely experimental and require primary tissue comparisons to be thoroughly vetted, especially for epigenetic modifications which can differ dramatically in vitro as compared to in vivo (Nestor et al., 2015). In recent years, however, technological advances have made it possible to examine epigenetic modifications from limited cell numbers, and in some cases within single cells. It is therefore now possible to investigate the epigenetic landscape in cells from in vivo inner ear tissues (or other related models) using a variety of different tools. Here, we provide an overview of several of these techniques, with a focus on those that have been validated for use with single cells or at least with limited cell numbers, and where applicable, provide examples for how these methods have been used by auditory and vestibular researchers. As one more point of caution, however, it is important to note that while these techniques have been demonstrated to work at the single cell level or with only hundreds or thousands of isolated cells in other systems, there are likely to be challenges in adopting some of these techniques to inner ear or related tissues. This is due not only to the small numbers of cells that can be isolated, but also the heterogeneity of the tissues and challenges faced in enriching for a single cell type. As there are stark differences in epigenetic features across different cell types, the importance of enriching for homogeneous cell populations cannot be understated. Data from many of these techniques with material isolated from heterogeneous populations of cells can be difficult to interpret as a method for assigning each detected modification to one cell type or another is currently lacking. Even homogeneous cell populations often do not reveal wholly conserved epigenetic features, but exhibit their own variability. Thus, it is critical for many of the experimental approaches outlined here that highly enriched cell populations be used, or, where possible, isolated single cells.
Methods for Detection of DNA Methylation
Perhaps the first technique to gain prominence for the investigation of epigenetic modifications is the use of sodium bisulfite to convert unmethylated cytosine residues at CpG dinucleotides to uracil residues (Frommer et al., 1992). Since methylated cytosines are protected from deamination by sodium bisulfite, direct comparison of bisulfite-treated and untreated DNA allows the localization of methylated cytosines. Depending on the scientific question and resources at hand, bisulfite treated DNA can be analyzed by PCR methods or by direct sequencing (Frommer et al., 1992; Li and Tollefsbol, 2011). PCR methods are generally useful when one gene or site within the genome, or at most a handful of sites, are of interest. Methylation-specific primers (MPs), designed to complement sites containing CpG dinucleotides, readily amplify methylated sites in bisulfite treated DNA (where CpG sequences are preserved), but are less efficient at amplifying these sites when they are unmethylated (i.e. unprotected from bisulfite and therefore converted into UpG sequences). Unmethylated-primers (UPs) are also generated and designed to cover the same sequences containing the CpG dinucleotides, but their sequences complement UpG residues at these sites. Thus comparisons using both sets of primers allows detection of cytosine methylation. For example, if the CpG sites are methylated, there should be robust amplification of bisulfite treated DNA with the MPs, but not with the UPs. If the site(s) of interest is unmethylated, then the UPs should give robust amplification and the MPs should be less efficient (Herman et al., 1996).
Nested methylation specific PCR (nested-MSP) is a modified version of this technique developed to detect methylation in small quantities of DNA (approximately 10ng). In nested-MSP, two rounds of PCR are used after bisulfite treatment. In the first round of PCR, primers that amplify independent of DNA methylation are designed to flank potential methylation sites in genes of interest. Unmethylated CpG residues will be converted to UpG residues by the bisulfite treatment and therefore amplified as TpG residues in the first round of PCR. These amplicons then undergo a second round of PCR with MPs and UPs that are nested within the first amplicon. Again there will be robust amplification of methylated CpGs with MPs, whereas the TpG residues generated from unmethylated CpGs will be amplified by the UPs (Scher et al., 2012).
Another PCR-based approach relies on thermal cycling coupled with a fluorometer to detect differences in melting temperature based on CpG vs. UpG content. For this method, primers are designed specifically to bracket, but not compliment, potential CpG sites that could be methylated. Following bisulfite treatment, methylated cytosine residues are amplified as such, whereas unmethylated cytosines, which were converted to uracils by the bisulfite treatment, are amplified as thymidines. The resulting amplicons will therefore have greater or lesser CpG content, respectively, which will result in different melting temperatures. The methylated samples with greater CpG content will melt at a higher temperature than the unmethylated samples with lesser CpG content (Guldberg et al., 2002). This type of analysis can be easily conducted using most commercially available qPCR cyclers and analysis software. Although we are not aware of any studies applying these techniques to single cells, amplification by PCR means that DNA methylation using bisulfite treatment can in theory be applied to the small numbers of cells typically found in auditory and vestibular organs, and should only require similar amounts of input as in typical real-time qPCR experiments.
Though simple, cost effective, and easy to carry out in most labs, PCR analyses of bisulfite treated DNA have two primary limitations. First, as the output of these experiments is binary (presence or absence of a band after a certain number of cycles, or higher vs. lower melting temperature), it can be difficult to determine how many methyl groups are present if more than one CpG dinucleotide is contained within the sequence being interrogated. Second, these methods are not suited to interrogate large portions of the genome, but rather are best for examining only one or a few DNA sequences at a time. The former limitation can be overcome by sequencing the PCR amplicons obtained in an approach termed bisulfite sequencing PCR, which allows the methylation status of individual CpG dinucleotides to be obtained (Li and Tollefsbol, 2011). However, to query large portions of the genome, one can employ whole-genome bisulfite sequencing (WGBS) or Bisulfite sequencing (BS-seq) which can most simply be described as the application of high-throughput sequencing methods to bisulfite treated DNA. In this method, DNA is isolated from the cells of interest, treated with bisulfite, and then cleaved into readable fragments which are ligated to adapters and sequenced (Clark et al., 2017, 1994; Cokus et al., 2008; Eckhardt et al., 2006; Smallwood et al., 2014). In this way, the whole genome can be sequenced and compared to reference genomes, as well as to sequences from untreated DNA. This allows for detection of (theoretically) all of the methylated sites within a cell’s genome. These methods are best performed in purified or homogeneous populations of cells as it is difficult to interpret or assign intermediate levels of methylation in heterogeneous populations.
Another approach that uses high-throughput sequencing to interrogate the whole genome for methylated cytosine residues is methylated DNA immunoprecipitation sequencing (MeDIP-seq) which relies on antibodies that recognize and bind specifically to methylated DNA. In this approach, DNA is extracted, fragmented, and immunoprecipitated using methylcytosine-specific antibodies. The antibodies are then removed by reverse crosslinking, the fragment ends are repaired and ligated to adapters, and then sequenced (Taiwo et al., 2012; Weber et al., 2005). If the starting quantity of DNA is a limiting factor (e.g. from a small number of cells obtained from a cochlea), MeDIP can also be followed by real-time qPCR for analysis to examine select genes rather than looking at the entire genome (Tsaliki et al., 2012).
Other options for interrogating global DNA methylation are amplification of intermethylated sites (AIMS) and comparative methylation hybridization (CMH), which rely on restriction endonucleases that are methylation sensitive (HpaII, SmaI) and insensitive (MspI, XmaI) to cut the DNA differentially. In AIMS, the digested DNA is ligated to an adapter, amplified by PCR, and fractionated on a gel for comparison between experimental and control samples (Frigola et al., 2002). While this approach generates a footprint for the overall methylation across the genome, it does not provide the locations or sequences of the methylated regions. However, partial sequences can be obtained by extracting bands of interest from the resulting gel and sequencing them. Also, while a methylated DNA footprint allows for quantification of the number of sites that are methylated, the frequency of methylation at a site across the cell population is difficult to quantitate or compare across groups with this method. In contrast to AIMS, CMH uses microarrays or high-throughput sequencing to assess the methylation status at sites recognized by the endonucleases and therefore can provide quantification of frequency within a cell population, as well as location and sequence information for these sites across the genome (Nelson, 2008). It is important to keep in mind that all high-throughput sequencing based approaches do not guarantee 100% coverage of the genome, particularly in samples with low input (e.g. single cells or low cell number) or in heterogeneous samples (e.g. anything other than single cells or clonal cells, the latter of which may still be heterogeneous in terms of DNA methylation). In addition, sites that are not uniformly methylated across most or all of the cells may not rise above the signal-to-noise ratio to be detected. Also, endonuclease based methods are limited by the sequence specificity of the endonucleases, cutting only at CCGG and/or CCCGGG sequences, but ignoring CpG sites not flanked by other C’s and G’s.
High performance liquid chromatography (HPLC) or liquid chromatography coupled with mass spectrometry (LC-MS) are also sensitive methods for measuring DNA methylation and may also be used to interrogate small numbers of cells (Armstrong et al., 2011; Ehrich et al., 2005; Lin et al., 2016). While these methods can provide an assessment of the global level of methylation in cell populations, they do not easily allow for localization of the methylated sites or quantification at single-loci resolution. In addition, HPLC and mass spectrometry require specialized equipment and technical expertise that may limit accessibility. In contrast, PCR based methods are more economical and more likely available in most labs that study the inner ear and related tissues. Table 1 summarizes the utility and caveats for each of these methods used to investigate DNA methylation.
Table 1:
Methods for Investigating DNA methylation
| Approach | Utility | Caveats | Examples from Inner Ear or Related Tissues |
|---|---|---|---|
| Bisulfite PCR with high resolution melt analysis | Rapid, cost-efficient method to determine if a specific region or multiple regions, of interest are methylated. | 1. Not useful for querying many regions of interest or for coverage of the whole genome. 2. Binary output (Y/N) may fail to reveal multiple methylation sites in a given region and does allow for comparison of methylation levels across experimental groups. |
N/A |
| Bisulfite sequencing PCR, Nested-MSP | Rapid, cost-efficient methods to determine if a specific region or multiple regions of interest are methylated or may have multiple sites of methylation. | 1. Not useful for querying many regions of interest or for coverage of the whole genome. |
Mutai et al., 2009 Wu et al., 2014 Lin et al., 2017 |
| EpiTyper (bisulfite PCR, transcription, MALDI-TOF MS) | Useful for determining the ratio of methylated vs unmethylated status of a particular region or several regions of the genome from a pool of cells or tissue of interest | 1. Not ideal for whole genome interrogation 2. Can be costly 3. Requires expertise and equipment for MALDI-TOF mass spectrometry |
Waldhaus et al., 2012 Xia et al., 2015 |
| BS-seq, WGBS | Useful for mapping methylation sites across the whole genome. If coupled with single cell isolation (scBS-seq) can detect monoallelic methylation. Allows for quantification and therefore comparison across experimental groups. |
1. Can be costly. 2. May require additional bioinformatic tools or other expertise with next generation sequencing and subsequent data analysis |
Yizhar-Barnea et al., 2018 |
| MeDIP-qPCR | Rapid, cost-effective method for determining whether there is methylated DNA in a specific genomic region or in a handful of regions of interest and how those levels may compare across experimental groups | 1. Not ideal for many regions of interest or whole genome interrogation 2. Does not provide absolute quantification, only relative comparison across groups |
Zhou and Hu, 2016 |
| MeDIP-seq | Useful for mapping methylation sites across the whole genome and providing quantitation and comparison across experimental groups. | 1. Can be costly. 2. May require additional bioinformatics and next generation sequencing expertise. 3. Single cell MeDIP is not yet well-established and cell sorting to obtain homogenous populations is therefore critical. |
N/A |
| AIMS | A versatile approach that offers similar benefits to BS-PCR, but can be used to provide information about global rather than local levels of DNA methylation. PCR amplification instead of sequencing results in cost savings. Gel extraction and sequencing of bands of interest can provide spatial localization of methylated sites of interest within the genome. DNA methylome footprints can be compared across samples. |
1. Use of sequence-specific endonucleases limits detection and biases the results toward sequences that are recognized by the endonuclease used. 2. Amplification rather than sequencing prevents localization of methylation sites across the genome. 3. Comparisons across groups are limited to quantitation of the number of sites that are overwhelmingly methylated within a sample. Frequencies of methylation at specific regions within a cell population cannot be determined or compared using this method. |
Mutai et al., 2009 |
| CMH | Provides site-specific methylation information across the genome. Use of microarrays instead of next-gen sequencing provides cost savings. Unlike AIMS, frequencies of methylation in a cell population can be determined and compared across samples. | 1. Genome coverage by microarray is limited compared to direct sequencing. 2. Sequence-specific endonucleases limit detection of all methylated sites similar to AIMS. |
N/A |
| LC-MS | Highly sensitive for detecting global levels of DNA methylation and does not require special enzymes or pretreatment. | 1. Can be costly. 2. Requires specialized equipment and expertise in mass spectrometry. 3. Does not provide location-specific information, only global levels of DNA methylation in a given sample. |
N/A |
| Genetic or pharmacological manipulation of DNMTs or TETs | Can uncover important processes and genes that are regulated by DNA methylation. Genetic manipulations can be designed to be cell-type specific obviating the need for cell sorting. | 1. Disruption of an enzyme may be fatal or, if not, may lead to complex and overlapping phenotypes due to targeting multiple loci. 2. Partiality of enzymes means coverage is not likely to be global. 3. Redundancy of enzymes means that genes that are regulated by methylation may be missed. 4. Context dependent divalence of enzymes to methylate or demethylate substrates may lead to further complexity of phenotypic results. 5. Potential off-target effects of pharmacological compounds may confound results. |
Chen et al., 2013 Zhou & Hu, 2015 Zhou & Hu, 2016 Roellig & Bronner, 2016 |
NOTE for Table 1: All of the techniques listed above with the exception of “genetic or pharmacological manipulation of DNMTs or TETs” are ideally performed using homogeneous or sorted cell populations or isolated single cells. Cellular heterogeneity can introduce a large degree of noise into the data and result in incorrect estimations of the frequencies and locations of DNA methylation in the genomes of cell types of interest.
Analysis of DNA Methylation in the Inner Ear (and Related Tissues)
Bisulfite sequencing PCR was used on DNA from cochlear extracts to examine methylation in two regions of interest in the promoter of Gap Junction Beta 2 or GJB2 which encodes Connexin 26 (Lin et al., 2017; Wu et al., 2014). Connexin 26 is a component of the gap junction channels found in SCs of the sensory epithelium and in fibrocytes of the spiral ligament and spiral limbus. These channels recycle potassium ions from the base of HCs to the endolymph and play an important role in homeostasis (Kikuchi et al., 2000). Mutations in GJB2 are found in up to 50% of patients with non-syndromic sensorineural hearing loss (Kenneson et al., 2002), making it the most common deafness-related gene. In a model of intrauterine hypoxia, Lin et al., (2017) used bisulfite sequencing PCR and found increased methylation of CpG sites in the GJB2 promoter of cochlear cells from 8 week old rats that were exposed to hypoxic conditions for 14 days in utero. This was accompanied by a ~40% decrease in Connexin 26 mRNA and protein expression, as well as significant hearing loss. A second study used a model of age-related hearing loss where rats were chronically treated with D-galactose which causes mitochondrial damage and increased formation of reactive oxygen species in the cochlea (Chen et al., 2011; Zhong et al., 2011). These rats had hypermethylation of CpG sites in the GJB2 promoter of cochlear cells at 10 months of age, as detected by bisulfite sequencing PCR, along with a 22–34% decrease in Connexin 26 mRNA and protein expression (Wu et al., 2014). Thus similar to congenital loss of function mutations in GJB2, hypermethylation of the GJB2 promoter can also decrease expression of Connexin 26 and cause hearing loss. Of note, both studies used DNA isolated from the whole cochlea and therefore it is unknown whether there are cell-type specific differences in the methylation of the GJB2 promoter between SCs and fibrocytes or among SC subtypes. Since bisulfite sequencing PCR was used, these studies could only assess select DNA sequences (i.e., two sites within the GJB2 promoter); therefore the methylation status of other genes in these models remains unknown.
A combination of nested-MSP and MeDIP-qPCR was used to investigate changes in the methylation of the promoter regions for three genes using a cell line created from mouse utricle sensory epithelial cells (MUCs) (Zhou and Hu, 2016). These cells expressed several HC-specific genes after treatment with 5-azacytidine, a compound which inhibits DNMT activity and therefore decreases DNA methylation (Chen et al., 2013). After bisulfite treatment, nested-MSP was performed on the promoter regions of Atoh1, Pou4f3, and Cdh1 (which encode the proteins ATOH1, POU4F3, and E-cadherin, respectively) and the brightness of the bands resulting from the PCR reactions using MPs vs. UPs were compared. MUCs treated with 5-azacytidine had significantly decreased amounts of methylated DNA in the promoter regions of Atoh1 and Pou4f3, with minimal change in the promoter of Cdh1. Using the more quantitative method of MeDIP-qPCR, the authors found a ~80% decrease in the methylation of Atoh1 and Pou4f3 promoters and no significant change in the methylation of the Cdh1 promoter. Experiments run in parallel found increases in the mRNA for each of these genes (Zhou and Hu, 2016). Similar to bisulfite sequencing PCR, both nested-MSP and MeDIP-qPCR used in this study only allowed the analysis of selected genes of interest. If the authors had instead used MeDIP-seq, they could have identified nearly all of the genes in MUCs with methylation changes after 5-azacytidine treatment in an unbiased manner. However as with any method that includes sequencing, MeDIP-seq is significantly more costly than MeDIP-qPCR and can require isolation and purification of greater starting quantities of DNA.
WGBS was used to generate genome-wide maps of DNA methylation at the single nucleotide resolution during key stages of inner ear maturation. Yizhar-Barnea et al., (2018) isolated DNA from the sensory epithelium of the mouse cochlea at embryonic day (E) 16.5, postnatal day (P) 0, and P22 and subjected the samples to WGBS. Though the DNA came from both sensory and nonsensory cells in the organ of Corti, and therefore neither the methylation patterns nor their dynamics could be assigned to specific cell types, several findings still emerged from the data. First, significantly more DNA methylation changes (both methylation and demethylation) occurred between P0 and P22 when cochlear maturation occurs, compared to E16.5 to P0 when cells are differentiating. Second, while assaying DNA methylation is often considered a means for identifying genomic regions where transcription is repressed, identifying unmethylated or low-methylated regions can suggest putative promoters and enhancers. From the data obtained in this study, many such potential transcription factor binding sites were identified, and, when mapped against available transcriptomic data, potential regulation sites for several genes known to be important in cochlear development and function were confirmed. In addition, novel findings included the identification of the transcription factor Bach2 and the HIF1 signaling pathway as potentially important factors in cochlear maturation. Finally, the authors used in silica tools to map the low methylated regions identified in the mouse genome to their orthologous sequence locations in the human genome. This mapping suggested numerous putative enhancers or promoters of genes that may be involved in human deafness, and the authors validated one of these sites using CRISPR-on technology (Cheng et al., 2013) which revealed that a low methylated region located between the known deafness genes GJB2 and GJB6 did not regulate the expression of the more proximal GJB2, but did have a pronounced effect on activating transcription of the more distal GJB6. This study very nicely highlights the wealth of data that can be obtained using WGBS, and the hypotheses that can be generated from such data, such as putative enhancer and promoter sequences, putative candidate genes that are likely to be expressed or repressed at different developmental stages or under different treatment conditions, and transcription factors and signaling pathways that may play a role in the cells and conditions being investigated. However, it is critical to remember that the predictions generated by WGBS and subsequent in silica analyses must be validated due not only to the size of the datasets generated, but also because the predictions may be flawed, and because not all unmethylated DNA is actively transcribed.
Mutai et al., (2009) used a modified version of AIMS combined with bisulfite sequencing PCR to investigate DNA methylation changes that occur in the sensory epithelium of rat cochleae before and after hearing onset. Using AIMS, comparison of the PCR products between P1 and P14 samples revealed a band specific to the P1 samples that was observed in repeated experiments. After this fragment was purified, re-amplified, and sequenced, it was identified as a potential regulatory element for the transcription factor Pou3f3. Bisulfite sequencing qPCR was then used to assess the methylation status of the Pou3f3 regulatory element at the two ages, where increased methylation was observed in the P14 samples along with a ~40% decrease in Pou3f3 mRNA. The use of AIMS in this study allowed a non-biased approach to identify Pou3f3 which had not previously been studied in the cochlea. The authors demonstrated that Pou3f3 is expressed in SCs and mesenchymal cells of the spiral ligament, spiral limbus, and Reissner’s membrane. Since no phenotype was observed in cochleae of Pou3f3 knock mice and the cells expressing Pou3f3 (i.e. cells of the greater epithelial ridge and mesenchyme) are known to decrease in number between P1 and P14, one caveat of this data which may affect its interpretation is that the P14 samples included more Pou3f3-negative cells than at P1. Since Pou3f3-negative cells are expected to have increased methylation of Pou3f3 (because this gene is not expressed), the methylation status of Pou3f3 in Pou3f3-positive cells at P14 remains unclear. This highlights one of the limitations of using heterogeneous tissue for epigenetic studies.
Two studies used bisulfite PCR followed by mass spectrometry to investigate changes in the methylation status of the regulatory elements for specific genes using inner ear tissue. Waldhaus et al., (2012) investigated methylation of promoter and enhancer regions of Sox2 in otospheres, derived from the postnatal cochlea. Sox2 is considered a pluripotency factor in embryonic and neural stem cells (Masui et al., 2007; Suh et al., 2007; Takahashi and Yamanaka, 2006). Yet in the inner ear it promotes the formation of prosensory progenitor cells (Kiernan et al., 2005) and neurons in the cochleovestibular ganglion (Steevens et al., 2017), and is required for the differentiation of HCs (Puligilla and Kelley, 2017) and SCs (Dabdoub et al., 2008) at later ages. Waldhaus et al., (2012) used a method called EpiTYPER where DNA is bisulfite treated, then amplified by PCR using primers that are amended with a T7 promoter. Following PCR, the reverse strands of the amplicons are transcribed using T7 polymerase and subsequently, the RNA molecules are cut into fragments using RNaseA. Fragments derived from methylated CpG islands will retain CG dinucleotide sequences in their RNA, whereas unmethylated CpG islands will be transcribed as CA nucleotides which have a 16 Da lower mass than CG nucleotides. The RNA fragments are analyzed with matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry which, for each cohort of RNA fragments, will yield two peaks 16 Da apart. The ratio of the signal intensities of these two peaks provides the ratio of methylated to unmethylated CpGs. Data from such an EpiTYPER experiment showed that the Sox2 promoter had similar levels of methylation in otospheres as in embryonic and neural stem cells, yet two enhancer regions, SRR1 and SRR2, had increased methylation in the otospheres compared to the other cell types. Next methylation of the otic-specific enhancers, NOP1 and NOP2, were examined in cells from E13.5, P4, and P21 cochleae where all four enhancer regions showed increased methylation with age. Interestingly when otospheres were grown under differentiating conditions or were treated with epidermal growth factor (EGF), the methylation of NOP1 and NOP2 was similar to P21 cochlear cells. In another study, Xia et al., (2015) used EpiTYPER to examine methylation of the Notch1 promoter in cochleae from neonatal and adult mice. Notch1 is a receptor that mediates Notch signaling, a pathway known to regulate HC differentiation and regeneration (Kelley, 2007; McGovern et al., 2018). Studies have shown that Notch signaling is downregulated in mouse cochleae during the first postnatal week through an unknown mechanism (Hartman et al., 2007; Maass et al., 2015; Murata et al., 2006). Xia et al., (2015) investigated methylation changes to the Notch1 promoter as a possible explanation for its decreased expression. DNA extracted from the auditory epithelium of P0, P4, P8, and P16 mice had a progressive increase in methylation of the Notch1 promoter with age, while Notch1 mRNA expression decreased over the same time period. The same experiments were performed in adult mice where HCs were killed by the aminoglycoside kanamycin. There was no change in the methylation of the Notch1 promoter in samples with HC damage compared to controls, yet there was a small increase in Notch1 mRNA expression. The authors concluded that Notch1 expression changes after HC damage are not regulated by DNA methylation. The combination of bisulfite PCR with mass spectrometry in these two examples allowed the measurement of methylation changes at specific DNA sequences which would not have been possible if mass spectrometry had been used alone. However mass spectrometry alone could have provided an assessment of global methylation changes across the genome.
Methods for Detection of Histone Modifications
Several methods have been developed for the study of histone modifications, some focusing on the proteins themselves and others on the DNA located in close proximity to histones that have been modified. LC-MS used alone can provide a snapshot of the relative frequency for all of the histone modifications (e.g. acetylation, methylation, phosphorylation, sumoylation, and ubiquitination) simultaneously. When coupled with peptide fragmentation or proteomics approaches, LC-MS can provide more precise information including the frequency of a specific modification at a single amino acid location on a histone protein (e.g. the percent of H3 proteins in a given sample that are acetylated at lysine 27), as well as grouping information (e.g. how many of those lysine 27 residues are mono- vs. di- vs. tri-methylated) (Önder et al., 2015). When combined with chromatin immunoprecipitation (ChIP) or chromatin proteomics approaches, regionally localized histones (i.e. those in a promoter or enhancer of interest) can also be assessed (Byrum et al., 2012; Waldrip et al., 2014; Wang et al., 2013).
For labs that do not have access to mass spectrometry equipment, reagents, and expertise, histone modifications can also be investigated by immunofluorescence, ChIP-qPCR, or ChIP-seq. For example, antibodies specific to methylation, dimethylation, or trimethylation of lysine 9 on histone H3 (H3K9me, H3K9me2, and H3K9me3, respectively) allow investigators not only the ability to visualize the relative prevalence of those histone modifications in cells and tissues of interest, but also to immunoprecipitate all of the histones that are methylated at H3K9 from a sample, and do so with specificity based on the number of methyl groups present. In recent years, there has been a vast expansion of commercially available antibodies against nearly all of the known modified residues on histone tails. These antibodies are useful for immunofluorescence to provide initial characterizations of the relative prevalence of specific histone modifications in a tissue (e.g. Layman et al., 2013). However, immunostaining as an approach for making direct comparisons of histone modifications across cell types, time points, or treatment conditions can be difficult because fluorescent intensities across images of immunostained samples can be variable and differences that are not global, but only occur on a limited number of histones per cell, may be too subtle to be detected. However, if single-cell resolution is not required, western blotting, ELISAs, or a bioanalyzer can be used to obtain relative quantification of the frequency of certain histone modifications from pools of cells or whole tissues (Dai et al., 2013; Wen et al., 2015). Additionally, with the increased use and improvement of super-resolution imaging techniques, image analyses of immunostained histone modifications may prove more useful in the near future. In the meanwhile, ChIP methods are the most common method for investigating histone modifications as they allow for more direct quantification. ChIP using an antibody against a specific histone modification of interest can be coupled with PCR or sequencing, or with mass spectrometry as aforementioned. In recent years, several modified ChIP protocols such as μChIP (Gilfillan et al., 2012) have been developed for use with small numbers of cells and in some cases, single cells (Rotem et al., 2015). In ChIP approaches, an antibody raised to bind selectively against a specific histone modification (e.g. anti-H3K9me2) is applied to nuclear lysates containing sheared DNA and DNA-binding proteins including the histones. After the antibodies bind, the antibody/histone complexes are immunoprecipitated, bringing the DNA in those nucleosomes along with it. The antibodies and histones are then removed by reverse cross-linking and primers specific to a region of interest can be used in PCR or qPCR to obtain relative quantification for the frequency of the histone modification of interest at the specific region of interest (Milne et al., 2009). To obtain a more global picture of the relative frequency of a particular modification (e.g. how much of the genome is associated with H3K9me2), the immunoprecipitated complexes can be quantified by mass spectrometry (Byrum et al., 2012; Waldrip et al., 2014; Wang et al., 2013). Finally, to determine not only how many histones in the genome are modified in a certain way, but also to identify the genomic sequences closest to those histones, the DNA obtained after reverse crosslinking can be adapted and sequenced using a next generation platform (i.e., ChIP-seq). In addition to its utility in identifying genomic regions associated with various histone marks, ChIP-seq using antibodies against certain well-described histone modifications can also, by proxy, point researchers in the direction of genes that may be activated or silenced under certain conditions (Klisch et al., 2011). For example, H3K4me3 is largely associated with active promoters of genes (Barski et al., 2007), so by performing ChIP-seq with an anti-H3K4me3 antibody, one can identify large cohorts of genes that are likely being actively transcribed. However, it is important to note that no single histone modification has been shown to correlate 100% with either activation or repression of gene expression, so ChIP-seq experiments like this are generally best for generating hypotheses about genes that may be activated or repressed with subsequent studies required to test those hypotheses directly. An additional approach that may be useful for interrogating histone modifications and the role(s) of histone modifying enzymes is DNA adenine methyltransferase identification sequencing (DamID-seq) (Steensel and Henikoff, 2000). By fusing histone writer or eraser proteins (e.g. a HAT or HDAC) to E coli DNA adenine methyltransferase, regions of the genome where the enzyme of interest is interacting with histones can be marked by the methylation of nearby adenines. The genomic positions of these sites can then be assayed by DpnI endonuclease treatment, which cleaves only at methylated adenines in GATC sequences (Kehat et al., 2011; Tolhuis et al., 2006). The resulting fragments can then be sequenced and mapped back to an intact reference genome sequence.
ChIP-seq is currently the most widely used method for assessing the prevalence of, as well as the genes most likely affected by, histone modifications. This wealth of information is certainly a strength of the approach and no doubt contributes to its swift and widespread adoption. However, it is important to consider that ChIP based approaches rely on antibodies which can have issues with specificity (e.g. off target binding) or sensitivity (i.e. incomplete coverage at rarer sites due to reduced binding kinetics). In contrast to this, the strength of DamID and LC-MS approaches are that they do not require antibodies and therefore may be more useful for targeted investigation of relatively rare histone modifications. In such instances, LC-MS would be useful for assessing the prevalence of a rare histone modification, while DamID-seq would be more useful for identifying genes affected by rare modifications. Table 2 summarizes the utility and caveats for each of these methods used to investigate histone modifications.
Table 2:
Methods for Investigating Histone Modifications
| Approach | Utility | Caveats | Examples from Inner Ear or Related Tissues |
|---|---|---|---|
| LC-MS | Can provide information about all of the modifications present on all of the histone proteins that are present in a sample. Can be used to compare global levels of histone modifications across experimental groups. | 1. Does not link individual histones and their modifications to specific locations in the genome. 2. Additional experiments are necessary to directly link changes in modifications to any phenotype. 3. Can be costly, require special equipment, and require additional expertise in mass spectrometry. |
N/A |
| Immunofluorescence, Western blotting | Economical, fast, and easily implemented in most laboratories. Can provide initial information and allow for correlation of various cellular processes with certain modifications that are present at high levels. | 1. Variability can result from differences in fixation, section thickness, age of sample, age of antibody, antibody promiscuity, and a number of other factors making direct quantification difficult and perhaps less reliable than other methods. 2. Requires finding and validating reliable antibodies. |
Mantela et al., 2005 Hansen et al., 2008 Weber et al., 2008 Chen et al. 2009 Sulg et al., 2010 Okano et al., 2011 Liu et al., 2012 Pan et al., 2013 Watanabe & Bloch 2013 Yu et al., 2013 Yue et al., 2013 Cox et al., 2014 Laos et al., 2014 Raft et al., 2014 Slattery et al., 2014 Walters et al., 2014 Huh et al., 2015 Urness et al., 2015 Chen et al., 2016 Li et al., 2017 |
| ChlP-PCR or ChIP qPCR | Cost effective method that allows for the determination of individual histone modification at small numbers of genomic sites of interest. If the PCR is quantitative, comparisons can be made across experimental groups. | 1. Is limited to the investigation of one, or at most a few, histone modifications of interest. 2. Is dependent on finding a reliable antibody against the modification of interest. 3. Only suitable for genes of interest as it does not provide genome-level information. |
Abdolazimi et al., 2016 Stojanova et al., 2016 Song et al. 2017 |
| ChlP-seq | Allows for the identification of all genomic regions that are bound to histones that have the modification of interest. Can be used to compare global levels of histone modifications across experimental groups. | 1. Is dependent on finding a reliable antibody with good binding kinetics. 2. Can be costly and may require additional bioinformatics and next generation sequencing expertise. |
Song et al., 2017 |
| DamlD-seq | Requires low input of DNA. Provides whole genome sequence data including all locations of the genome where the histone modifying enzyme of interest was in close proximity to the DNA. Depending on the approach used, it is possible to generate cell type-specific expression of the adenine methyl-transferase-conjugated enzyme thus limiting the need for cell sorting. | 1. Requires knowledge of the histone modifying enzyme of interest. 2. Requires genetic editing approaches to fuse adenine methyltransferase to the histone modifying enzyme of interest. 3. If the histone modifying enzyme has multiple roles involving DNA, the resulting data may include sites not related to the histone modification of interest. 4. May require additional bioinformatics and next generation sequencing expertise |
N/A |
| Genetic or pharmacological manipulation of writer and eraser enzymes | Can uncover important processes and genes that are regulated by a histone modification of interest (or by a set of histone modifications). Genetic manipulation can be designed to be cell type specific obviating the need for cell sorting. | 1. Overlap of function across a variety of histone writing and erasing enzymes as well as promiscuous activity of these enzymes on other proteins may yield complex phenotypes and can make it difficult to tie phenotypes directly to the histone modification being tested. 2. Potential off-target effects of pharmacological compounds may confound results. |
Drottar et al., 2006 Stojanova et al., 2009 He et al., 2013 Yu et al., 2013 He et al., 2015 Layman et al., 2015 Li et al., 2015 Wang et al., 2015 Chen et al., 2016 He et al., 2016 Stojanova et al., 2016 Yang et al., 2017 |
NOTE for Table 2: All of the techniques listed above with the exception of “DamiID-seq” and “Genetic or pharmacological manipulation of writer and eraser enzymes” are ideally performed with sorted or homogeneous populations of cells or isolated single cells. Cellular heterogeneity can introduce a large degree of noise into the data resulting in incorrect estimations of the frequency and locations of histone modifications in the cell types of interest.
Analysis of Histone Modifications in the Inner Ear (and Related Tissues)
There are several recent studies where immunofluorescence was used to study histone modifications in the inner ear and related tissues. Two studies used antibodies generated against acetylated histone residues and image analysis to measure the effect of gentamycin-induced or noise-induced HC damage on histone acetylation (Chen et al., 2009, 2016). Using cochlear explants treated with gentamycin, Chen et al., (2009) found a decrease in H2A, H2B, H3, and H4 acetylation over time as HCs began to die. Yet there was no change in histone acetylation in neighboring Hensen cells, a subtype of SCs. In adult mice exposed to high intensity noise that induces permanent threshold shifts, there was a decrease in the H3K9ac marks in outer HCs located in the basal turn of the cochlea and in marginal cells of the stria, yet no change in H3K9ac was detected in the outer HCs of apical and middle turns or in spiral ganglion neurons (SGNs) (Chen et al., 2016). Another study examined changes in both acetylation and methylation of H3K9 by immunostaining and confocal microscopy. In this report, H3K9ac was readily detectable in the SGN and in the organs of Corti from young mice, but not detectable in aged mice. By contrast, H3K9me2 was detectable in cells in aged cochleae, particularly in the basal turns where HC loss was evident, but H3K9me2 was not readily detected in young cochleae (Watanabe and Bloch, 2013). These results led the authors to speculate that more genes may be epigenetically silenced with increasing age and that increased levels of H3K9me2 may presage HC death. Consistent with this idea, Yu et al., (2013) also using immunostaining and western blotting, demonstrated increased levels of H3K9me2 in response to a number of ototoxic insults (cisplatin, copper, ultraviolet light, and neomycin) in organotypic cochlear cultures. The benefit of studies such as these is that they can demonstrate readily detectable levels of several acetylated and methylated histone residues in numerous cell types in the cochlea, and provide initial comparisons of histone acetylation and methylation status between control and ototoxic conditions and/or between neighboring cells within the same samples. However, as mentioned above, the caveats to immunofluorescent approaches such as this, is that they are only semi-quantitative, and do not reveal the specific regions of the genome where histones are modified. Nor can they provide high resolution information about the dynamics of acetylation/deacetylation or methylation/demethylation across the entire population of histones in a given cell or pool of cells. Specifically, studies of this type only allow for the determination of a net effect (e.g. overall decrease of H3K9ac immunoreactivity), but it is likely that, within any given cell, histone proteins in different parts of the genome are being modified differently (e.g. acetylation of H3K9 in some nucleosomes and deacetylation at other locations). Therefore, immunostaining and western blotting reveal only large net effects, but do not provide information about which genes are being silenced and which genes may be being activated in response to a particular stimulus.
There are, however, instances where immunostaining of histone modifications can reveal relatively straightforward information regarding critical cellular processes. This is particularly true for certain histone residues that become widely phosphorylated during proliferation or cell death. For example, phosphorylation of serine 10 on histone H3 (H3S10ph) has been shown to be important for M phase of the cell cycle, and the detection of H3S10ph by immunostaining can be used to identify cells that were in M phase at the time of fixation. Indeed, several inner ear or related papers have used anti-H3S10ph antibodies in this manner (Cox et al., 2014; Huh et al., 2015; Li et al., 2017; Liu et al., 2012; Mantela et al., 2005; Okano et al., 2011; Pan et al., 2013; Raft et al., 2014; Sulg et al., 2010; Urness et al., 2015; Walters et al., 2014; Weber et al., 2008). While useful, it is important to note that H3S10ph has also been shown to play a role in DNA damage/repair and cell death, where, for example, H3S10ph may be increased in response to cisplatin (Park and Kim, 2012), a chemotherapeutic agent known to kill HCs. This is an important factor to consider when using H3S10ph as a marker for cell cycle, and other methods should be used in conjunction to confirm proliferation and/or rule out cell death. Another important phosphorylated site that may be detectable using immunostaining is phosphorylation of serine 139 on the H2A variant, H2AX. Indeed, H2AXS139 phosphorylation (aka γ-H2AX) occurs in a rapid and robust manner near double strand breaks in the DNA, and is essential for recruiting repair complexes, as well as arresting cell cycle progression (Podhorecka et al., 2010). Since double strand breaks are a hallmark of apoptosis (Didenko and Hornsby, 1996), immunostaining for γ-H2AX is often used as a marker for apoptotic cells, and several investigations of inner ear and related tissues have utilized anti-γ-H2AX staining for this purpose (Hansen et al., 2008; Laos et al., 2014; Slattery et al., 2014; Sulg et al., 2010; Yue et al., 2013).
ChIP-qPCR was recently used to probe histone modifications in the regulatory regions of Atoh1 during embryonic and postnatal cochlear maturation. Atoh1 is a transcription factor that is necessary for HC differentiation (Bermingham, 1999; Woods et al., 2004). Its expression follows a bell-shaped curve where prosensory cells initiate expression between E13.5–14.5, followed by downregulation after HC differentiation between E17.5 and P6 (Driver et al., 2013; Kelley, 2006; Maass et al., 2015; Woods et al., 2004). Stojanova et al., (2016) used fluorescent activated cell sorting (FACS) to purify either prosensory cells, HCs, or SCs and then performed μCHIP-qPCR with H3K27me3, H3K4me3, H3K9me3, or H3K9ac antibodies using ~25,000 cells per reaction. Three regions in the Atoh1 locus contained H3K27me3 and H3K4me3 bivalent marks in prosensory cells from E14.5 cochlea. These histone modifications are known to maintain genes in a “poised”, but not expressed state (Bernstein et al., 2006). In contrast, in HCs at E17.5, H3K27me3 and H3K4me3 bivalent marks were dramatically reduced. In addition, there was an increase in the H3K9ac marks on the Atoh1 locus at E17.5 which generally promotes gene expression (Karmodiya et al., 2012). These epigenetic marks corresponded with a ~50 fold increase in Atoh1 expression between E14.5 and E17.5 and SCs at P1 maintained the H3K27me3 and H3K4me3 bivalent marks observed in prosensory cells. When comparing FACS-purified HCs from E17.5 and P6 cochlea, a time period when Atoh1 expression is downregulated, H3K9ac marks decreased and there was an increase in H3K9me3, an epigenetic modification associated with gene repression (Rea et al., 2000). Another study (Abdolazimi et al., 2016) used the same methods to investigate histone acetylation changes in the Atoh1 locus after neonatal cochlear explants were treated with the gamma-secretase inhibitor DAPT, which stimulates the conversion of SCs into HCs and activates Atoh1 expression (Doetzlhofer et al., 2009; Golden et al., 2015; Korrapati et al., 2013; W. Li et al., 2015; Maass et al., 2015). Abdolazimi et al., (2016) found that 24 hours after DAPT treatment, FACS-purified SCs had increased H3K9ac marks in the Atoh1 locus. The combination of FACS and μCHIP-qPCR used in these studies provided a focused analysis of the time-dependent and cell-type specific changes to the epigenetic landscape of Atoh1. However only one gene was investigated here and changes in histone modifications may be occurring in many other genes as well, which could have been elucidated if ChIP-seq or DamID-seq were used.
In a recent paper, ChIP-seq and ChIP-qPCR were utilized to identify candidate genes important in the differentiation of immortalized multipotent otic progenitor (iMOP) cells into SGN-like cells in culture (Song et al., 2017). iMOP cells are an immortalized cell line derived from Sox2-expressing cells from ~E12 mouse otocysts that have been transduced with retrovirus to express c-Myc and are maintained in a proliferative state by supplementation with basic fibroblast growth factor (bFGF). Upon removal of bFGF, iMOP cells can differentiate into HC-like cells, SC-like cells, and SGN-like cells (Kwan et al., 2015). Interestingly, when the proneural gene Neurog1 is ectopically expressed in proliferating iMOP cells, it promotes further proliferation, but in differentiating iMOP cells, Neurog1 promotes differentiation into SGN-like cells which are non-mitotic. To identify the genes involved in this switch from proliferation to differentiation in iMOP cells, Song et al (2017) performed ChIP-seq using antibodies against H3K4me3, H3K27me3, and RNA polymerase II. Antibodies against H3K4me3 and RNA polymerase II should immunoprecipitate promoter regions in activated genes, while antibodies against H3K27me3 are more likely to precipitate regions that are repressed (Barski et al., 2007). Since iMOP cells in culture are a heterogeneous mixture of cells that may be either proliferative, non-differentiating or differentiating, non-proliferative, the authors hypothesized that regions of DNA immunoprecipitated by both the marks for transcriptional activation and repression would be critical in the shift from proliferation to differentiation and therefore critical to the process of SGN-like cell differentiation. The ChIP-seq data identified Cdk2 and Neurod1 as potential regulators of iMOP cell proliferation and differentiation, respectively. ChIP-qPCR using antibodies against H3K9ac or H3K9me3 and primers targeted to the promoters of Cdk2 and Neurod1 further supported the hypothesis that Cdk2 promoters were more likely to be active in proliferating cells, while Neurod1 promoters were more likely to be active in differentiated cells. Specifically, amplification of H3K9ac immunoprecipitated DNA yielded more Cdk2 promoter sequences in proliferating iMOP cells as compared to differentiated SGN-like cells, and this pattern was reversed when Neurod1 promoter sequences were amplified. When antibodies against the repressive mark H3K9me3 were used to immunoprecipitate the DNA, more Cdk2 sequences were amplified in the differentiated SGN-like cells than in the proliferating iMOP cells, while more Neurod1 promoter sequences were amplified in the proliferating iMOPs than in the SGN-like cells. These experiments, combined with others in the paper, strongly suggest that Neurog1 can promote Cdk2 to enhance proliferation in the bFGF condition, and promote Neurod1 to enhance SGN-like cell differentiation when bFGF is absent. Furthermore, the ChIP-qPCR data suggest that it may be the status of H3K9 at the Cdk2 and Neurod1 promoters that acts as the switch to determine whether Neurog1 will promote Cdk2 and proliferation or Neurod1 and differentiation. The use of ChIP-seq in this study highlights its utility for identifying promoters and genes that are differentially regulated under diverse conditions, while ChIP-qPCR was used to support and extend these results.
Methods for the Manipulation of Epigenetic Writers and Erasers
While detection of changes in DNA methylation or histone modifications during development, after a drug treatment, or during disease progression is informative, these epigenetic changes can also be intentionally altered using pharmacological agents or genetic tools. For example DNMT inhibitors such as 5-azacytidine and 5-aza-2-deoycytidine have been used with MUC cells where they caused decreased methylation in promoter regions of the genes investigated (Zhou and Hu, 2016, 2015). HAT inhibitors such as curcumin and HDAC inhibitors such as valproic acid, trichostatin A, sodium butyrate, and suberoylanilide hydroxamic acid (SAHA) will prevent or remove histone acetylation, respectively. Curcumin was used in cochlear explant cultures where it reduced the numbers of HCs and decreased expression of Atoh1 mRNA, which corresponded with decreased H3K9ac marks in the Atoh1 locus (Stojanova et al., 2016). Many studies have shown that HDAC inhibitors can have a protective effect against noise-induced (Chen et al., 2016; Yang et al., 2017) or ototoxic drug-induced (Chen et al., 2009; Drottar et al., 2006; Layman et al., 2015; Wang et al., 2015; Yang et al., 2017) HC damage and hearing loss. These pharmacological agents have a broad and global effect, altering epigenetic changes across the genome.
Yu et al., (2013) used the compounds BIX01294 or UNC0638 to pharmacologically inhibit G9a/GLP methyltransferase. This in turn reduced H3K9me2 levels in HCs in cochlear explants and, in vivo, promoted HC survival against neomycin-induced ototoxicity. Similarly, pharmacological inhibition of lysine specific demethylase 1 (LSD1) using the monoamine oxidase inhibitor 2-PCPA has been shown to cause increased histone methylation and apoptosis in zebrafish neuromasts during development (He et al., 2013). Interestingly, however, two similar studies using pharmacological inhibitors of LSD1 suggested that histone methylation may actually prevent apoptotic death of HCs in response to neomycin treatment (He et al., 2015) and of SGNs in response to cisplatin treatment (Li et al., 2015). Genetic disruption studies, as well as more detailed mapping of histone methylation in response to toxic insults, are likely needed to determine whether there may be any generalizable aspects of these histone marks on cell survival, or if the effects of methylation at H3K4 and/or H3K9 are regulating different populations of genes in different cell types. In addition to influencing cell survival, histone methylation has been implicated in proliferation where LSD1 activity appears to be necessary for proliferation during development and HC regeneration in zebrafish neuromasts. Again, pharmacological inhibition of LSD1 by 2-PCPA was used and resulted in an increase in H3K4me2 immunostaining which was correlated with decreased proliferation and decreased HC regeneration (He et al., 2016, 2013).
Others have opted for a genetic approach. For example, morpholino based knockdown of Dnmt3a, a DNA methylating enzyme, resulted in a significant reduction in the size of the otic placode and reduced expression of several otic-specific genes (Pax2, Gbx2, Sox10, and Soho1) in developing chicken embryos. In contrast, the expression patterns of several other otic-specific genes (Otx2, Sox2, and Sox3) were unaffected by Dnmt3a knockdown (Roellig and Bronner, 2016). Interestingly, this report suggests that DNA methylation is necessary for the activation of certain genes. While several possible mechanisms could explain this such as: 1) repression of repressor proteins leading to activation of downstream targets, 2) repression of genes in cells of one domain causing activation of genes in cells in an adjoining domain via cell signaling, or 3) DNA methylation may not always lead to transcriptional repression, but may block access of repressive complexes to DNA thereby promoting gene expression. These data are again an important reminder that one cannot presume to ascribe an all-up or all-down characteristic to any epigenetic modification.
Methods to Assess Chromatin Conformation
In epigenetic studies, sometimes it is not the modifications to the DNA or the histones that are important, but rather a determination of how accessible the DNA in a particular region(s) of interest may be at different times or under different experimental conditions. Several techniques have been developed that allow for the assessment of chromatin structure. Perhaps the most prominent method is the assay for transposase accessible chromatin sequencing, or ATAC-seq (Buenrostro et al., 2013). ATAC-seq works by treating DNA with an engineered transposase enzyme that has high cutting and ligating activity on accessible DNA; that is, DNA which is not protected by being tightly condensed around histone complexes. Treatment with the hyperactive Tn5 transposase not only cuts accessible DNA, but also simultaneously ligates adapters for high throughput sequencing. Sequenced reads from single cells then provide a map of regions where the chromatin is in an open conformation. When using pools of cells, sequence read numbers positively correlate with the probability of a site being open across the cell population. However, it is again important to note that data from heterogeneous cell populations will not be as robust as data from sorted or clonal cell populations. To avert this potential problem, ATAC-seq can be conducted on single cells (Buenrostro et al., 2015), suggesting that it is amenable to the study of auditory and vestibular tissues. Indeed, ATAC-seq has already been used to study developing mouse otocysts and cochleae (Gálvez et al., 2017). Although the authors were only interested in the regulation of a single gene, Atoh1, ATAC-seq provided whole genome maps that identified the regions that were more accessible at E10.5 versus E14.5. These two timepoints were chosen because at E10.5 Atoh1 is repressed by Neurog1 during neurogenesis and at E14.5 there is autoactivation of Atoh1 by ATOH1 binding to its 3’ enhancer during HC differentiation. ATAC-seq was also run on E14.5 cochleae that were treated with the gamma-secretase inhibitor LY411575 which causes upregulation of Atoh1 via the inhibition of Notch signaling. While there did not appear to be any differences in accessibility of the Atoh1 promoter or enhancer regions across these three different samples, the ATAC-seq data did reveal that the A region of the 3’ enhancer remained inaccessible in all three groups, while the B region of the enhancer was open, suggesting that the B region of the enhancer is critical for regulating Atoh1 expression in the developing cochlea. Subsequent validation experiments showed that the enhancer B region is the primary driver for Atoh1 positive feedback and a critical site for Neurog1-mediated repression of Atoh1.
DNase-seq was a predecessor to ATAC-seq and is still a viable alternative to ATAC-seq, or can be conducted in parallel to ATAC-seq for increased robustness of data. DNase-seq operates on a similar premise in which condensed chromatin is protected from nuclease activity and therefore accessible chromatin can be digested into small fragments that are isolated and sequenced (Jin et al., 2015; Song and Crawford, 2010). Chromosome conformation capture methods (3C, 4C, 5C, Hi-C, and capture-C) are additional useful approaches for assaying chromatin accessibility (Belton et al., 2012; Dekker et al., 2002). Although these methods are primarily used for detecting regions of chromatin that interact across distances (e.g. distal promoters and enhancers), they can also reveal regions where there is a large degree of condensed DNA or DNA brought into close proximity by histone modifications. These methods work by crosslinking closely appositioned DNA/protein complexes with formaldehyde. The DNA is then digested by restriction enzymes into fragments that undergo proximity ligation (so that non-complexed DNA is not ligated), and reverse cross-linking then linearizes the resulting hybrid DNA. In chromosome conformation capture (3C, see Dekker et al., 2002), ligations of close proximity DNA are measured by qPCR where a single set of primers can be designed to interrogate whether two non-sequential regions of the genome interact, or a matrix of primer pairs across numerous qPCR reactions can be used to interrogate multiples of such hypothetical close proximity relationships. In chromosome conformation capture-on-chip (4C, see Simonis et al., 2006), microarrays or next-gen sequencing (4C-seq) are used rather than qPCR to assess close proximity interactions of DNA across the genome. To accomplish this, the 3C protocol is followed up to and including ligation and reverse crosslinking, after which an additional restriction digest is performed and then an additional ligation to create circular DNA constructs. Primer and PCR amplification is again employed, but in this approach, the circular DNA allows for both primers to be designed to be complementary to sequences in only one of the DNA fragments (rather than requiring one primer to bind in each of the two ligated fragments). The amplicons are then identified and quantified by microarray or sequencing. With the 4C approach it is therefore possible to query all of the regions of DNA that interact with one region of interest. In chromosome conformation capture carbon copy (5C, see Dostie et al., 2006), the 3C library is generated by fixation, digestion, ligation, and reverse crosslinking, and then a “carbon copy” is made by annealing forward and reverse oriented oligonucleotides that are designed to contain sequences corresponding to the 3’ ends of the restriction fragments and with universal (T7 or T3) tails for amplification. When these 5C oligos anneal next to each other they can be ligated by Taq ligase to create unique oligonucleotides containing sequences that map to both interacting pieces. These new fragments (the 5C library) can then be amplified by PCR using universal primers and can then be assessed by microarray or sequencing. Due to the requirements for specially designed oligos, 5C does not provide complete coverage of the genome, but it does allow for a less biased, higher throughput approach than 3C for interrogating many regions of the DNA and many of the interacting regions. Hi-C (see Lieberman-Aiden et al., 2009) was the first “all versus all” approach derived from 3C and works by introducing biotinylated nucleotides into the 5’ overhangs of the digested DNA prior to the proximity ligation step during the construction of the 3C library. After this, the DNA is subjected to reverse cross-linking, purification, and shearing and then a biotin pull down enriches for ligated fragments which are then sequenced. Hi-C thereby provides an unbiased approach to interrogate the whole genome for interacting regions of DNA and the data can be used to generate three dimensional maps of chromatin structure. In a related protocol called ChIA-PET (chromatin interaction analysis by paired end tag sequencing, (see Fullwood et al., 2009) regions of DNA that are in close proximity, as well as DNA-binding proteins are fixed with formaldehyde, the chromatin is sheared and antibodies against proteins of interest (e.g. histone subunits) are immunoprecipitated to enrich for DNA associated with those proteins and the DNA associated with that DNA. Closely interacting DNA then undergoes proximity ligation and paired-end-tagging followed by sequencing and data analysis. Although the authors are not aware of studies with inner ear or related tissues that used these approaches, single cell Hi-C (Nagano et al., 2015) and Hi-ChIP, a low sample input version of ChIA-PET (Mumbach et al., 2016), have been established and may prove useful for such experiments. Table 3 summarizes the utility and caveats for each of these methods used to investigate chromatin structure.
Table 3:
Methods for Investigating Chromatin Structure
| Approach | Utility | Caveats | Examples from Inner Ear or Related Tissues |
|---|---|---|---|
| ATAC-seq | Provides genome-level data revealing the probabilities of finding any given region in an open or closed state across a cell population. | 1. Can be costly 2. May require additional bioinformatics and next generation sequencing expertise. |
Galvez et al., 2017 |
| DNase-seq | Provides genome-level data revealing the probabilities of finding any given region in an open or closed state across a cell population. | 1. Can be technically challenging as it requires determination of optimal DNase concentration and duration of treatment which is variable across sample types. 2. May require pooling of samples to achieve the required numbers of cells. 3. Can be costly 4. May require additional bioinformatics and next generation sequencing expertise. |
N/A |
| Chromatin capture techniques (3C, 4C, 5C, Hi-C) | Provides structural information about chromatin from cells or tissues of interest. Is particularly useful for discovery of distal promoters and enhancers. Can provide indirect information about the level to which DNA is condensed or open. |
1. Can be costly 2. May require additional expertise, including bioinformatic assistance to map ligated sequences to two or more disparate locations in the genome. 3. ATAC-seq and DNase-seq provide more direct measures of chromatin accessibility. |
N/A |
| ChIA-PET, Hi-ChIP | Highly useful for investigating the role of specific DNA interacting proteins and their effects on chromatin structure. | 1. Can be costly 2. May require additional expertise, including bioinformatic assistance to map ligated sequences to two or more disparate locations in the genome. 3. May require pooling of cells as the ChIP step reduces the amount of input material obtained per cell. |
N/A |
NOTE for Table 3: All of the techniques listed above are ideally performed with sorted or homogeneous populations of cells or isolated single cells. Cellular heterogeneity can introduce a large degree of noise into the data resulting in incorrect estimations of the frequencies of chromatin interactions or the degree to which DNA in certain regions is accessible in a given cell type.
Conclusion
This review is far from exhaustive, both in regard to the tools and approaches presented, as well as to the epigenetics studies that have been undertaken in the inner ear and related tissues. For example, there are many protein complexes that regulate gene expression that are recruited to histones by the modifications that have been discussed. One such example CHD7, a chromodomain helicase that is recruited to enhancer regions of DNA and positively correlates with transcription factor recruitment and gene activation, is recruited to the DNA by H3K4me (Schnetz et al., 2010). As mutations in CHD7 are a known cause of CHARGE syndrome, which can result in hearing loss and vestibular dysfunction, there are a number of articles that have examined the role of CHD7 in the development and function of auditory and vestibular tissues (Choo et al., 2017; Green et al., 2014; Micucci et al., 2014; Yao et al., 2018). Review of these and other articles pertaining to histone related proteins are beyond the scope of this already extensive review. However, the pharmacological and genetic methods described above for the manipulation of histone writers and erasers are similar to the types of approaches one would most likely use to pursue questions about the functions of histone related proteins like CHD7. Furthermore, acetylation, methylation, and phosphorylation represent only a fraction of the types of modifications that can occur on histone residues. Indeed, histones, like many other proteins, can be ubiquinated (Weake and Workman, 2008), sumoylated (Shiio and Eisenman, 2003), glycosylated (Sakabe et al., 2010), ribosylated (Hottiger, 2015), biotinylated, citrullinated, crotonylated, succinylated, and the list goes on (Xu et al., 2014). This suggests that there is still much left to explore regarding the roles of histone modifications, not only in the inner ear, but more broadly as well. Many of the tools outlined above for the study of histone acetylation and methylation can be repurposed for the study of these modifications as well. Genetic and pharmacological manipulations of the writer and eraser enzymes may also provide valuable insights into the functions of various histone modifications in many cellular processes important in the development, proliferation, differentiation, survival, and function of the cells in the inner ear and related tissues.
Highlights.
Low cell numbers or other challenges have hindered inner ear epigenetic research
Recent methodological advances provide the means for overcoming such challenges
DNA methylation, histone modifications, and chromatin accessibility can be assayed
Methods and examples of epigenetic studies of hearing and balance are presented
Acknowledgements
This work was supported in part by the National Institutes of Health (R01 DC014441 to BC and R01 DC016365 to BW), the Office of the Assistant Secretary of Defense for Health Affairs (W81XWH-15-1-0475 to BC), the Office of Naval Research (N00014-18-1-2716 to BW), and the American Otological Society (Research Grant to BW).
Abbreviations:
- AIMS
amplification of intermethylated sites
- ATAC-seq
assay for transposase accessible chromatin sequencing
- bFGF
basic fibroblast growth factor
- BS-seq
bisulfite sequencing
- ChIA-PET
chromatin interaction analysis by paired end tag sequencing
- ChIP
chromatin immunoprecipitation
- CMH
comparative methylation hybridization
- CpG
cytosine residue followed by a guanine residue
- DamID-seq
DNA adenine methyltransferase identification sequencing
- DNMT
DNA methyltransferase
- E
embryonic day
- EGF
epidermal growth factor
- FACS
fluorescent activated cell sorting
- HAT
histone acetyltransferase
- HDAC
histone deacetylate
- HCs
hair cells
- HPLC
high performance liquid chromatography
- iMOP
immortalized multipotent otic progenitor
- JmjC
jumonji C
- LC-MS
liquid chromatography coupled with mass spectrometry
- LSD1
lysine specific demethylase 1
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- MeDIP-seq
methylated DNA immunoprecipitation sequencing
- MP
methylation-specific primer
- MUCs
mouse utricle sensory epithelial cells
- nested-MSP
nested methylation specific PCR
- P
postnatal day
- PRMT
protein arginine N-methyltransferase
- SAHA
suberoylanilide hydroxamic acid
- SCs
supporting cells
- SGN
spiral ganglion neuron
- TET
ten-eleven translocation
- UP
unmethylated primers
- UpG
uracil residue followed by a guanine residue
- WGBS
whole-genome bisulfite sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
Brandon C. Cox, PhD is a consultant for Turner Scientific, LLC and Otonomy, Inc.
References
- Abdolazimi Y, Stojanova Z, Segil N, 2016. Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter. Development 143, 841–850. 10.1242/dev.129320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong KM, Bermingham EN, Bassett SA, Treloar BP, Roy NC, Barnett MP, 2011. Global DNA methylation measurement by HPLC using low amounts of DNA. Biotechnol. J 6, 113–117. 10.1002/biot.201000267 [DOI] [PubMed] [Google Scholar]
- Baek SH, 2011. When signaling kinases meet histones and histone modifiers in the nucleus. Mol. Cell 42, 274–84. 10.1016/j.molcel.2011.03.022 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K, 2007. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 129, 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Belton J-M, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J, 2012. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods 58, 268–276. 10.1016/j.ymeth.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, 1999. Math1: An Essential Gene for the Generation of Inner Ear Hair Cells. Science. 284, 1837–1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES, 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–26. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bird A, 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP, 1999. Methylation-induced repression--belts, braces, and chromatin. Cell 99, 451–4. [DOI] [PubMed] [Google Scholar]
- Biswas S, Rao CM, 2018. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol 837, 8–24. 10.1016/j.ejphar.2018.08.021 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ, 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–8. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ, 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum SD, Raman A, Taverna SD, Tackett AJ, 2012. ChAP-MS: A Method for Identification of Proteins and Histone Posttranslational Modifications at a Single Genomic Locus. Cell Rep. 2, 198–205. 10.1016/j.celrep.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SM, Gozani O, 2016. Nonhistone Lysine Methylation in the Regulation of Cancer Pathways. Cold Spring Harb. Perspect. Med 6 10.1101/cshperspect.a026435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DT, Chai R, DiMarco R, Heilshorn SC, Cheng AG, 2015. Protein-engineered hydrogel encapsulation for 3-D culture of murine cochlea. Otol. Neurotol 36, 531–8. 10.1097/MAO.0000000000000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W, D’Amore PA, 2008. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 19, 111–20. 10.1016/j.cytogfr.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zhong Y, Peng W, Sun Y, Hu Y, Yang Y, Kong W, 2011. Increased mitochondrial DNA damage and decreased base excision repair in the auditory cortex of D-galactose-induced aging rats. Mol. Biol. Rep 38, 3635–42. 10.1007/s11033-010-0476-5 [DOI] [PubMed] [Google Scholar]
- Chen C, Pan D, Deng A-M, Huang F, Sun B-L, Yang R-G, 2013. DNA methyltransferases 1 and 3B are required for hepatitis C virus infection in cell culture. Virology 441, 57–65. 10.1016/j.virol.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Chen F-Q, Schacht J, Sha S-H, 2009. Aminoglycoside-induced histone deacetylation and hair cell death in the mouse cochlea. J. Neurochem 108, 1226–1236. 10.1111/j.1471-4159.2009.05871.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hill K, Sha SH, 2016. Inhibitors of Histone Deacetylases Attenuate Noise-Induced Hearing Loss. J. Assoc. Res. Otolaryngol 17, 289–302. 10.1007/s10162-016-0567-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R, 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171. 10.1038/cr.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo DI, Tawfik KO, Martin DM, Raphael Y, 2017. Inner ear manifestations in CHARGE: Abnormalities, treatments, animal models, and progress toward treatments in auditory and vestibular structures. Am. J. Med. Genet. C. Semin. Med. Genet 175, 439–449. 10.1002/ajmg.c.31587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M, 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Smallwood SA, Lee HJ, Krueger F, Reik W, Kelsey G, 2017. Genome-wide base-resolution mapping of DNA methylation in single cells using single-cell bisulfite sequencing (scBS-seq). Nat. Protoc 12, 534–547. 10.1038/nprot.2016.187 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE, 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–9. 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen D-H, Chalasani K, Steigelman KA, Fang J, Cheng AG, Zuo J, 2014. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829. 10.1242/dev.103036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones J, 2008. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 105, 18396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Giardina C, Rasmussen TP, 2013. Quantitation of Nucleosome Acetylation and Other Histone Posttranslational Modifications Using Microscale NU-ELISA. Methods Mol. Biol 981, 167–176. 10.1007/978-1-62703-305-3_13 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N, 2002. Capturing chromosome conformation. Science 295, 1306–11. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Didenko VV, Hornsby PJ, 1996. Presence of double-strand breaks with single-base 3’ overhangs in cells undergoing apoptosis but not necrosis. J. Cell Biol 135, 1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A, Avraham KB, 2017. Insights into inner ear-specific gene regulation: Epigenetics and non-coding RNAs in inner ear development and regeneration. Semin. Cell Dev. Biol 65, 69–79. 10.1016/j.semcdb.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N, 2009. Hey2 Regulation by FGF Provides a Notch-Independent Mechanism for Maintaining Pillar Cell Fate in the Organ of Corti. Dev. Cell 16, 58–69. 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J, 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–309. 10.1101/gr.5571506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver EC, Sillers L, Coate TM, Rose MF, Kelley MW, 2013. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol 376, 86–98. 10.1016/j.ydbio.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drottar M, Liberman MC, Ratan RR, Roberson DW, 2006. The histone deacetylase inhibitor sodium butyrate protects against cisplatin-induced hearing loss in guinea pigs. Laryngoscope 116, 292–6. 10.1097/01.mlg.0000197630.85208.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB, 2002. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–9. 10.1093/embo-reports/kvf053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S, 2006. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet 38, 1378–1385. 10.1038/ng1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret G, Frankenreiter M, 1977. Quantitative analysis of cochlear structures in the house mouse in relation to mechanisms of acoustical information processing. J. Comp. Physiol 122, 65–85. 10.1007/BF00611249 [DOI] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D, 2005. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A 102, 15785–15790. 10.1073/pnas.0507816102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, 2007. Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–40. 10.1038/nature05919 [DOI] [PubMed] [Google Scholar]
- Frigola J, Ribas M, Risques R-A, Peinado MA, 2002. Methylome profiling of cancer cells by amplification of inter-methylated sites (AIMS). Nucleic Acids Res. 30, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL, 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A 89, 1827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed Y. Bin, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren W-J, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung W-K, Liu ET, Wei C-L, Cheung E, Ruan Y, 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64. 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez H, Tena JJ, Giraldez F, Abelló G, 2017. The Repression of Atoh1 by Neurogenin1 during Inner Ear Development. Front. Mol. Neurosci 10, 321 10.3389/fnmol.2017.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Hughes T, Sheng Y, Hjorthaug HS, Straub T, Gervin K, Harris JR, Undlien DE, Lyle R, 2012. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics 13, 645 10.1186/1471-2164-13-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden EJ, Benito-Gonzalez A, Doetzlhofer A, 2015. The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea. Proc. Natl. Acad. Sci. U.S.A 112, E3864–E3873. 10.1073/pnas.1501077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green GE, Huq FS, Emery SB, Mukherji SK, Martin DM, 2014. CHD7 mutations and CHARGE syndrome in semicircular canal dysplasia. Otol. Neurotol 35, 1466–70. 10.1097/MAO.0000000000000260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y, 2012. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet 13, 343–357. 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldberg P, Worm J, Grønbaek K, 2002. Profiling DNA methylation by melting analysis. Methods 27, 121–7. [DOI] [PubMed] [Google Scholar]
- Hansen MR, Clark JJ, Gantz BJ, Goswami PC, 2008. Effects of ErbB2 signaling on the response of vestibular schwannoma cells to gamma-irradiation. Laryngoscope 118, 1023–30. 10.1097/MLG.0b013e318163f920 [DOI] [PubMed] [Google Scholar]
- Hartman BH, Hayashi T, Nelson BR, Bermingham-McDonogh O, Reh TA, 2007. Dll3 is expressed in developing hair cells in the mammalian cochlea. Dev. Dyn 236, 2875–83. 10.1002/dvdy.21307 [DOI] [PubMed] [Google Scholar]
- He Y, Tang D, Cai C, Chai R, Li H, 2016. LSD1 is Required for Hair Cell Regeneration in Zebrafish. Mol. Neurobiol 53, 2421–2434. 10.1007/s12035-015-9206-2 [DOI] [PubMed] [Google Scholar]
- He Y, Yu H, Cai C, Sun S, Chai R, Li H, 2015. Inhibition of H3K4me2 Demethylation Protects Auditory Hair Cells from Neomycin-Induced Apoptosis. Mol. Neurobiol 52, 196–205. 10.1007/s12035-014-8841-3 [DOI] [PubMed] [Google Scholar]
- He Y, Yu H, Sun S, Wang Y, Liu I, Chen Z, Li H, 2013. Trans-2-phenylcyclopropylamine regulates zebrafish lateral line neuromast development mediated by depression of LSD1 activity. Int. J. Dev. Biol 57, 365–73. 10.1387/ijdb.120227hl [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB, 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U. S. A 93, 9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, 2015. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem 84, 227–63. 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- Huh S-H, Warchol ME, Ornitz DM, 2015. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. Elife 4 10.7554/eLife.05921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, Levens D, Zhao K, 2015. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–6. 10.1038/nature15740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L, 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424 10.1186/1471-2164-13-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I, Accornero F, Aronow BJ, Molkentin JD, 2011. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J. Cell Biol 193, 21–29. 10.1083/jcb.201101046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW, 2007. Cellular commitment and differentiation in the organ of Corti. Int. J. Dev. Biol 51, 571–583. 10.1387/ijdb.072388mk [DOI] [PubMed] [Google Scholar]
- Kelley MW, 2006. Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci 7, 837–849. 10.1038/nrn1987 [DOI] [PubMed] [Google Scholar]
- Kenneson A, Van Naarden Braun K, Boyle C, 2002. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet. Med 4, 258–74. 10.1097/00125817-200207000-00004 [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KKH, Tang ASP, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KSE, 2005. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–1035. 10.1038/nature03487 [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Kimura R, Paul D, Takasaka T, 2000. Gap junction systems in the mammalian cochlea. Brain Res. 32, 163–6. [DOI] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY, 2011. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc. Natl. Acad. Sci. U. S. A 108, 3288–3293. 10.1073/pnas.1100230108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, Hashino E, 2014. 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat. Protoc 9, 1229–1244. 10.1038/nprot.2014.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y, 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–9. 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrapati S, Roux I, Glowatzki E, Doetzlhofer A, 2013. Notch Signaling Limits Supporting Cell Plasticity in the Hair Cell-Damaged Early Postnatal Murine Cochlea. PLoS One 8, e73276 10.1371/journal.pone.0073276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Shen J, Corey DP, 2015. C-MYC transcriptionally amplifies SOX2 target genes to regulate self-renewal in multipotent otic progenitor cells. Stem cell reports 4, 47–60. 10.1016/j.stemcr.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laos M, Anttonen T, Kirjavainen A, Hällström TA, Laiho M, Pirvola U, af Hällström T, Laiho M, Pirvola U, 2014. DNA damage signaling regulates age-dependent proliferative capacity of quiescent inner ear supporting cells. Aging. 6, 496–510. 10.18632/aging.100668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman WS, Sauceda MA, Zuo J, 2013. Epigenetic alterations by NuRD and PRC2 in the neonatal mouse cochlea. Hear. Res 304, 167–178. 10.1016/j.heares.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman WS, Williams DM, Dearman J. a, Sauceda M. a, Zuo J, 2015. Histone deacetylase inhibition protects hearing against acute ototoxicity by activating the Nf-κB pathway. Cell Death Discov. 1, 1–16. 10.1038/cddiscovery.2015.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, He Y, Sun S, Cai C, Li H, 2015. Lysine-specific demethylase 1 inhibitors protect cochlear spiral ganglion neurons against cisplatin-induced damage. Neuroreport 26, 539–547. 10.1097/WNR.0000000000000386 [DOI] [PubMed] [Google Scholar]
- Li W, Wu J, Yang J, Sun S, Chai R, Chen Z-Y, Li H, 2015. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci U S A 112, 166–171. 10.1073/pnas.1415901112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tollefsbol TO, 2011. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol. Biol 791, 11–21. 10.1007/978-1-61779-316-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Watanabe K, Fujioka M, Ogawa K, 2017. Characterization of slow-cycling cells in the mouse cochlear lateral wall. PLoS One 12, e0179293 10.1371/journal.pone.0179293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J, 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–93. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Huang H, Lv G, Xu X, Lin W, Xu X, Cheng J, Zheng M, 2017. Chronic prenatal hypoxia impairs cochlear development, a mechanism involving connexin26 expression and promoter methylation. Int. J. Mol. Med 41, 852–858. 10.3892/ijmm.2017.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X-C, Zhang T, Liu L, Tang H, Yu R-Q, Jiang J-H, 2016. Mass Spectrometry Based Ultrasensitive DNA Methylation Profiling Using Target Fragmentation Assay. Anal. Chem 88, 1083–1087. 10.1021/acs.analchem.5b04247 [DOI] [PubMed] [Google Scholar]
- Liu Z, Walters B, Owen T, Brimble MA, Steigelman KA, Zhang L, Lagarde MMM, Valentine MB, Yu Y, Cox BC, Zuo J, 2012. Regulation of p27 Kip1 by Sox2 maintains quiescence of inner pillar cells in the murine auditory sensory epithelium. J. Neurosci 32, 10530–10540. 10.1523/JNEUROSCI.0686-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass JC, Gu R, Basch ML, Waldhaus J, Lopez EM, Xia A, Oghalai JS, Heller S, Groves AK, 2015. Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front. Cell. Neurosci 9, 110 10.3389/fncel.2015.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantela J, Jiang Z, Ylikoski J, Fritzsch B, Zacksenhaus E, Pirvola U, 2005. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development 132, 2377–88. 10.1242/dev.01834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MSH, Niwa H, 2007. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol 9, 625–35. 10.1038/ncb1589 [DOI] [PubMed] [Google Scholar]
- McGovern MM, Zhou L, Randle MR, Cox BC, 2018. Spontaneous hair cell regeneration is prevented by increased notch signaling in supporting cells. Front. Cell. Neurosci 12, 120 10.3389/fncel.2018.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micucci JA, Layman WS, Hurd EA, Sperry ED, Frank SF, Durham MA, Swiderski DL, Skidmore JM, Scacheri PC, Raphael Y, Martin DM, 2014. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum. Mol. Genet 23, 434–448. 10.1093/hmg/ddt435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Zhao K, Hess JL, 2009. Chromatin immunoprecipitation (ChIP) for analysis of histone modifications and chromatin-associated proteins. Methods Mol. Biol 538, 409–23. 10.1007/978-1-59745-418-6_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY, 2016. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922. 10.1038/nmeth.3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Tokunaga A, Okano H, Kubo T, 2006. Mapping of Notch activation during cochlear development in mice: Implications for determination of prosensory domain and cell fate diversification. J. Comp. Neurol 497, 502–18. 10.1002/cne.20997 [DOI] [PubMed] [Google Scholar]
- Mutai H, Nagashima R, Sugitani Y, Noda T, Fujii M, Matsunaga T, 2009. Expression of Pou3f3 / Brn-1 and its genomic methylation in developing auditory epithelium. Dev. Neurobiol 69, 913–930. 10.1002/dneu.20746 [DOI] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, Tanay A, Fraser P, 2015. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc 10, 1986–2003. 10.1038/nprot.2015.127 [DOI] [PubMed] [Google Scholar]
- Nelson S, 2008. Comparative Methylation Hybridization. Nat. Educ 1, 55. [Google Scholar]
- Nestor CE, Ottaviano R, Reinhardt D, Cruickshanks HA, Mjoseng HK, McPherson RC, Lentini A, Thomson JP, Dunican DS, Pennings S, Anderton SM, Benson M, Meehan RR, 2015. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 16, 11 10.1186/s13059-014-0576-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T, Xuan S, Kelley MW, 2011. Insulin-Like Growth Factor Signaling Regulates the Timing of Sensory Cell Differentiation in the Mouse Cochlea. J. Neurosci 31, 18104–18118. 10.1523/JNEUROSCI.3619-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önder Ö, Sidoli S, Carroll M, Garcia BA, 2015. Progress in epigenetic histone modification analysis by mass spectrometry for clinical investigations. Expert Rev. Proteomics 12, 499–517. 10.1586/14789450.2015.1084231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Jin Y, Chen J, Rottier RJ, Steel KP, Kiernan AE, 2013. Ectopic Expression of Activated Notch or SOX2 Reveals Similar and Unique Roles in the Development of the Sensory Cell Progenitors in the Mammalian Inner Ear. J. Neurosci 33, 16146–16157. 10.1523/JNEUROSCI.3150-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C-H, Kim K-T, 2012. Apoptotic phosphorylation of histone H3 on Ser-10 by protein kinase Cδ. PLoS One 7, e44307 10.1371/journal.pone.0044307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podhorecka M, Skladanowski A, Bozko P, 2010. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J. Nucleic Acids 2010, 1–9. 10.4061/2010/920161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Kelley MW, 2017. Dual role for Sox2 in specification of sensory competence and regulation of Atoh1 function. Dev. Neurobiol 77, 3–13. 10.1002/dneu.22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raft S, Andrade LR, Shao D, Akiyama H, Henkemeyer M, Wu DK, 2014. Ephrin-B2 governs morphogenesis of endolymphatic sac and duct epithelia in the mouse inner ear. Dev. Biol 390, 51–67. 10.1016/j.ydbio.2014.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T, 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–9. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Roellig D, Bronner ME, 2016. The epigenetic modifier DNMT3A is necessary for proper otic placode formation. Dev. Biol 411, 294–300. 10.1016/j.ydbio.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto D, Avvakumov N, Côté J, 2012. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7, 1098–108. 10.4161/epi.21975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE, 2015. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol 33, 1165–1172. 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW, 2010. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U. S. A 107, 19915–20. 10.1073/pnas.1009023107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher MB, Elbaum MB, Mogilevkin Y, Hilbert DW, Mydlo JH, Sidi AA, Adelson ME, Mordechai E, Trama JP, 2012. Detecting DNA methylation of the BCL2, CDKN2A and NID2 genes in urine using a nested methylation specific polymerase chain reaction assay to predict bladder cancer. J. Urol 188, 2101–7. 10.1016/j.juro.2012.08.015 [DOI] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, LaFramboise T, Tesar P, Wei C-L, Scacheri PC, 2010. CHD7 Targets Active Gene Enhancer Elements to Modulate ES Cell-Specific Gene Expression. PLoS Genet. 6, e1001023 10.1371/journal.pgen.1001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Côté J, Chua KF, Gozani O, 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–9. 10.1038/nature04835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN, 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A 100, 13225–30. 10.1073/pnas.1735528100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W, 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet 38, 1348–54. 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Slattery EL, Oshima K, Heller S, Warchol ME, 2014. Cisplatin exposure damages resident stem cells of the mammalian inner Ear. Dev. Dyn 243, 1328–1337. 10.1002/dvdy.24150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G, 2014. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820. 10.1038/nmeth.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE, 2010. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010, pdb.prot5384. 10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Jadali A, Fritzsch B, Kwan KY, 2017. NEUROG1 Regulates CDK2 to Promote Proliferation in Otic Progenitors. Stem Cell Reports 9, 1516–1529. 10.1016/j.stemcr.2017.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S, 2000. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat. Biotechnol 18, 424–428. 10.1038/74487 [DOI] [PubMed] [Google Scholar]
- Steevens AR, Sookiasian DL, Glatzer JC, Kiernan AE, 2017. SOX2 is required for inner ear neurogenesis. Sci. Rep 7, 4086 10.1038/s41598-017-04315-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanova ZP, Kwan T, Segil N, 2016. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development 143, 3529–36. 10.1242/dev.137976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH, 2007. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell 1, 515–28. 10.1016/j.stem.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulg M, Kirjavainen A, Pajusola K, Bueler H, Ylikoski J, Laiho M, Pirvola U, 2010. Differential sensitivity of the inner ear sensory cell populations to forced cell cycle re-entry and p53 induction. J. Neurochem 112, 1513–26. 10.1111/j.1471-4159.2009.06563.x [DOI] [PubMed] [Google Scholar]
- Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, Beck S, Butcher LM, 2012. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc 7, 617–636. 10.1038/nprot.2012.012 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–76. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M, 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet 38, 694–9. 10.1038/ng1792 [DOI] [PubMed] [Google Scholar]
- Tsaliki E, Papageorgiou EA, Spyrou C, Koumbaris G, Kypri E, Kyriakou S, Sotiriou C, Touvana E, Keravnou A, Karagrigoriou A, Lamnissou K, Velissariou V, Patsalis PC, 2012. MeDIP real-time qPCR of maternal peripheral blood reliably identifies trisomy 21. Prenat. Diagn 32, 996–1001. 10.1002/pd.3947 [DOI] [PubMed] [Google Scholar]
- Urness LD, Wang X, Shibata S, Ohyama T, Mansour SL, 2015. Fgf10 is required for specification of non-sensory regions of the cochlear epithelium. Dev. Biol 400, 59–71. 10.1016/j.ydbio.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhaus J, Cimerman J, Gohlke H, Ehrich M, Müller M, Löwenheim H, 2012. Stemness of the organ of Corti relates to the epigenetic status of Sox2 enhancers. PLoS One 7, e36066 10.1371/journal.pone.0036066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip ZJ, Byrum SD, Storey AJ, Gao J, Byrd AK, Mackintosh SG, Wahls WP, Taverna SD, Raney KD, Tackett AJ, 2014. A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics 9, 1207–11. 10.4161/epi.29919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Diao S, Zheng F, Walters BJ, Layman WS, Zuo J, 2016. Pseudo-immortalization of postnatal cochlear progenitor cells yields a scalable cell line capable of transcriptionally regulating mature hair cell genes. Sci. Rep 5, 17792 10.1038/srep17792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Liu Z, Crabtree M, Coak E, Cox BC, Zuo J, 2014. Auditory hair cell-specific deletion of p27Kip1in postnatal mice promotes cell-autonomous generation of new hair cells and normal hearing. J. Neurosci 34, 15751–63. 10.1523/JNEUROSCI.3200-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, Britton L-MP, Elledge SJ, Kharchenko PV, Garcia BA, Kuroda MI, 2013. Chromatin proteins captured by ChIP–mass spectrometry are linked to dosage compensation in Drosophila. Nat. Struct. Mol. Biol 20, 202–209. 10.1038/nsmb.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Y, Chen X, Zhang P, Shi Z, Wen L, Qiu J, Chen F, 2015. Histone deacetylase inhibitor sodium butyrate attenuates gentamicin-induced hearing loss in vivo. Am. J. Otolaryngol 36, 242–248. 10.1016/j.amjoto.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Watanabe K-I, Bloch W, 2013. Histone methylation and acetylation indicates epigenetic change in the aged cochlea of mice. Eur. Arch. Oto-rhino-laryngology 270, 1823–30. 10.1007/s00405-012-2222-1 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL, 2008. Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–63. 10.1016/j.molcel.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D, 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet 37, 853–862. 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- Weber T, Corbett MK, Chow LML, Valentine MB, Baker SJ, Zuo J, 2008. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc. Natl. Acad. Sci. U.S.A 105, 781–785. 10.1073/pnas.0708061105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L-T, Wang J, Wang Y, Chen F-Q, 2015. Association between histone deacetylases and the loss of cochlear hair cells: Role of the former in noise-induced hearing loss. Int. J. Mol. Med 36, 534–540. 10.3892/ijmm.2015.2236 [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW, 2004. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci 7, 1310–8. 10.1038/nn1349 [DOI] [PubMed] [Google Scholar]
- Wu X, Wang Y, Sun Y, Chen S, Zhang S, Shen L, Huang X, Lin X, Kong W, 2014. Reduced expression of Connexin26 and its DNA promoter hypermethylation in the inner ear of mimetic aging rats induced by d-galactose. Biochem. Biophys. Res. Commun 452, 340–346. 10.1016/j.bbrc.2014.08.063 [DOI] [PubMed] [Google Scholar]
- Xia Y, Cao X, Xue X, Feng Z, Fan Q, Zheng Y, Feng C, Xu H, Xia C, Cheng Y, 2015. Development of hair cells in inner ear is associated with expression and promoter methylation of Notch-1 in postnatal mice. Int. J. Clin. Exp. Med 8, 15542–8. [PMC free article] [PubMed] [Google Scholar]
- Xu Y-M, Du J-Y, Lau ATY, 2014. Posttranslational modifications of human histone H3: an update. Proteomics 14, 2047–60. 10.1002/pmic.201300435 [DOI] [PubMed] [Google Scholar]
- Yang C-H, Liu Z, Dong D, Schacht J, Arya D, Sha S-H, 2017. Histone Deacetylase Inhibitors Are Protective in Acute but Not in Chronic Models of Ototoxicity. Front. Cell. Neurosci 11, 315 10.3389/fncel.2017.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-H, Xie J, Liu K, Peng Z, Guo J-Y, Yu S-K, Wang G-P, Gong S-S, 2017. The histone deacetylase inhibitor sodium butyrate protects against noise-induced hearing loss in Guinea pigs. Neurosci. Lett 660, 140–146. 10.1016/j.neulet.2017.09.036 [DOI] [PubMed] [Google Scholar]
- Yao H, Hill SF, Skidmore JM, Sperry ED, Swiderski DL, Sanchez GJ, Bartels CF, Raphael Y, Scacheri PC, Iwase S, Martin DM, 2018. CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development. JCI Insight 3, pii: 97440 10.1172/jci.insight.97440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar-Barnea O, Valensisi C, Jayavelu ND, Kishore K, Andrus C, Koffler-Brill T, Ushakov K, Perl K, Noy Y, Bhonker Y, Pelizzola M, Hawkins RD, Avraham KB, 2018. DNA methylation dynamics during embryonic development and postnatal maturation of the mouse auditory sensory epithelium. Sci. Rep 8, 17348 10.1038/s41598-018-35587-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lin Q, Wang Y, He Y, Fu S, Jiang H, Yu Y, Sun S, Chen Y, Shou J, Li H, 2013. Inhibition of H3K9 methyltransferases G9a/GLP prevents ototoxicity and ongoing hair cell death. Cell Death Dis. 4, e506–e506. 10.1038/cddis.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WY, Clark JJ, Telisak M, Hansen MR, 2013. Inhibition of c-Jun N-terminal kinase activity enhances vestibular schwannoma cell sensitivity to gamma irradiation. Neurosurgery 73, 506–16. 10.1227/01.neu.0000431483.10031.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Hu Y-J, Yang Y, Peng W, Sun Y, Chen B, Huang X, Kong W-J, 2011. Contribution of common deletion to total deletion burden in mitochondrial DNA from inner ear of d-galactose-induced aging rats. Mutat. Res 712, 11–9. 10.1016/j.mrfmmm.2011.03.013 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Hu Z, 2016. Epigenetic DNA Demethylation Causes Inner Ear Stem Cell Differentiation into Hair Cell-Like Cells. Front. Cell. Neurosci 10, 185 10.3389/fncel.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hu Z, 2015. Genome-wide demethylation by 5-aza-2’-deoxycytidine alters the cell fate of stem/progenitor cells. Stem Cell Rev. 11, 87–95. 10.1007/s12015-014-9542-z [DOI] [PMC free article] [PubMed] [Google Scholar]