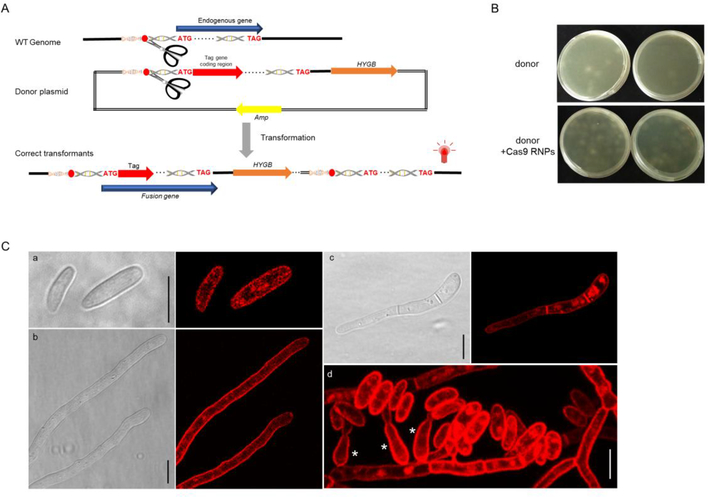
Fig. 4.
FoSSO2 gene tagging with a N-terminal mCherry fluorescent marker based on the HITI strategy. (A) The scheme for the HITI strategy of the N-terminal tagging of the FoSSO2 gene; a ~800 bp homologous sequence containing the sgRNA cleavage site in front of the FoSSO2 gene start codon was cloned and ligated to the mCherry marker coding sequence, the FoSSO2 endogenous gene, the 3′-UTR region of the FoSSO2 gene, and the hygromycin resistance cassette. During transformation, the entire plasmid will be inserted at the sgRNA cleavage site generating the N-terminal mCherry reporter for FoSSO2. (B) Comparison of the transformation efficiency with or without Cas9 RNPs for the FoSSO2 N-terminal tagging. (C) mCherry-FoSso2 subcellular localization in different fungal structures. a: conidia, b: fungal tips, c: a germling, d: fungal conidiation, *: phialides Scale bars, 10 μm.