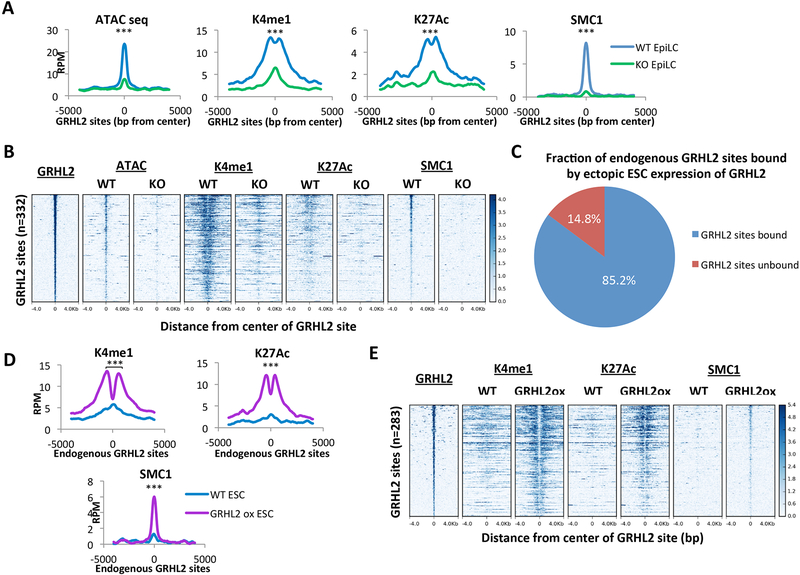
Figure 3. GRHL2 is necessary and sufficient for full enhancer activation.
A) Metagene analysis of average signal across GRHL2 sites in GRHL2 knockout and wildtype (V6.5) EpiLCs. All plots are shown for an 8000bp window centered at GRHL2 binding sites. RPM = reads per million. K4me1 p = 8.79167E-50; K27ac p = 1.77148E-52; SMC1 p = 3.08123E-40; ATAC: p = 7.13881E-72 (paired t-test). B) Heatmaps at GRHL2 sites for enhancer marks in GRHL2 knockout and wildtype (V6.5) EpiLCs. All plots are shown for an 8kb window centered at GRHL2 binding sites. Sites are shown in descending order based on mean GRHL2 ChIP signal intensity. C) Fraction of endogenous GRHL2 binding sites in EpiLCs that are bound by GRHL2 overexpression in ESCs (GRHL2 sites bound). D) Metagene analysis of average signal across endogenous GRHL2 sites that are ectopically bound in GRHL2 overexpressing ESCs in untargeted (WT) cells and dox-inducible GRHL2 ESCs, both treated with doxycycline. RPM = reads per million. K4me1 p = 1.56194E-45; K27ac p = 3.69415E-24; SMC1 p = 1.0148E-34 (paired t-test). E) Heatmaps showing all individual loci in (D). All plots are shown for an 8kb window centered at GRHL2 binding sites. Sites are shown in descending order based on mean GRHL2 ChIP signal intensity.