Figure 6: Functional validation of enhancer switching at GRHL2 target genes during the ESC to EpiLC transition.
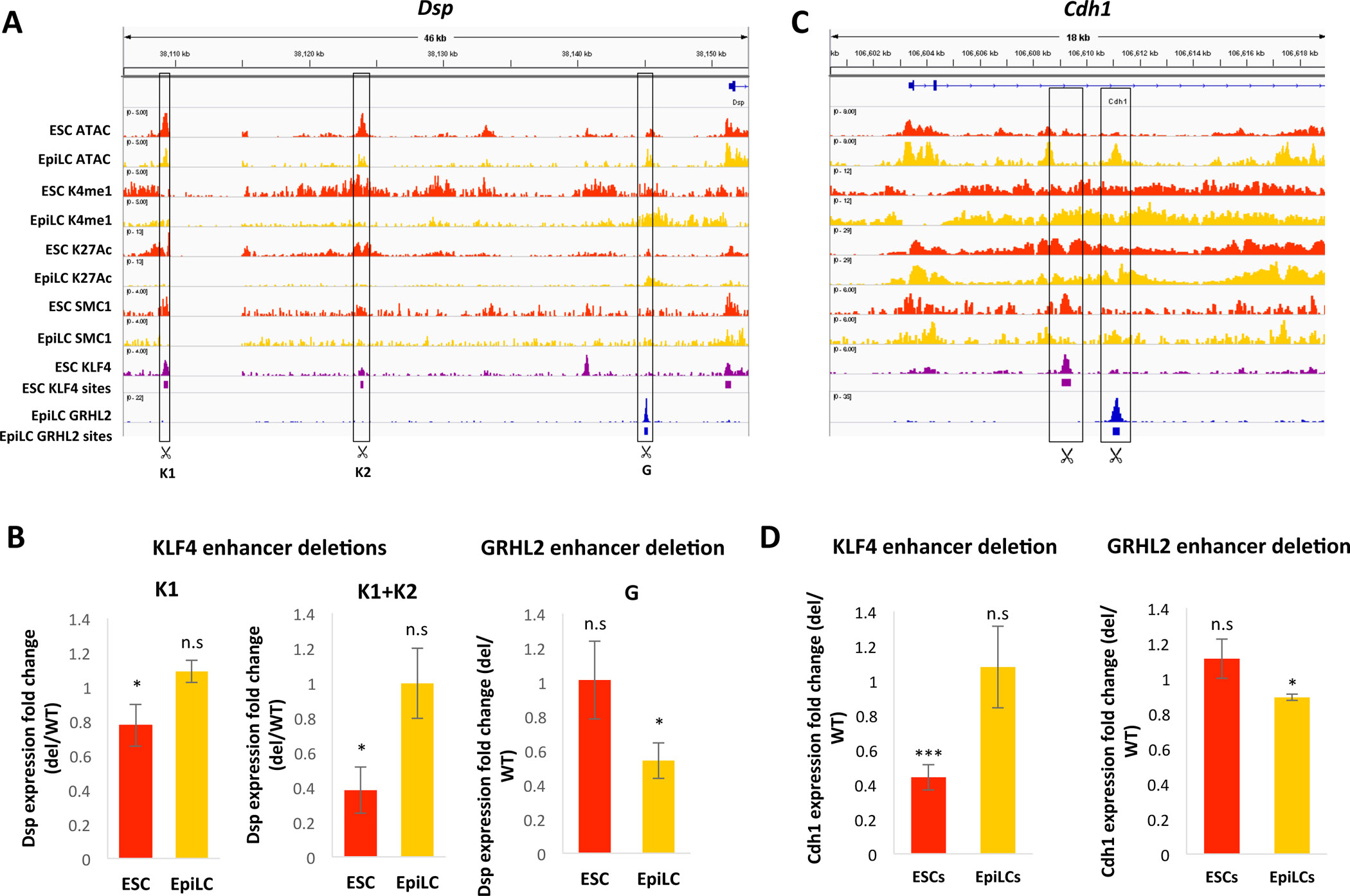
A) ChIP-seq and ATAC-seq tracks in WT ESCs and EpiLCs at the Dsp locus, with significant KLF4 and GRHL2 binding sites indicated by purple or blue bars, respectively. KLF4 and GRHL2 sites deleted using CRISPR are indicated. B) Fold change in Dsp expression with deletion of the indicated KLF4 enhancers (K1 alone, or K1 and K2) or GRHL2 enhancer in ESCs and EpiLCs. Astericks (*) indicate p < 0.05 by Student’s t-test. C) Same as in (A) but for the Cdh1 locus. D) Fold change in Cdh1 expression with deletion of the nearby KLF4-bound enhancer (left) or GRHL2-bound enhancer (right) in ESCs and EpiLCs. * indicates p < 0.05 and *** indicates p< 0.005 by Student’s t-test.
