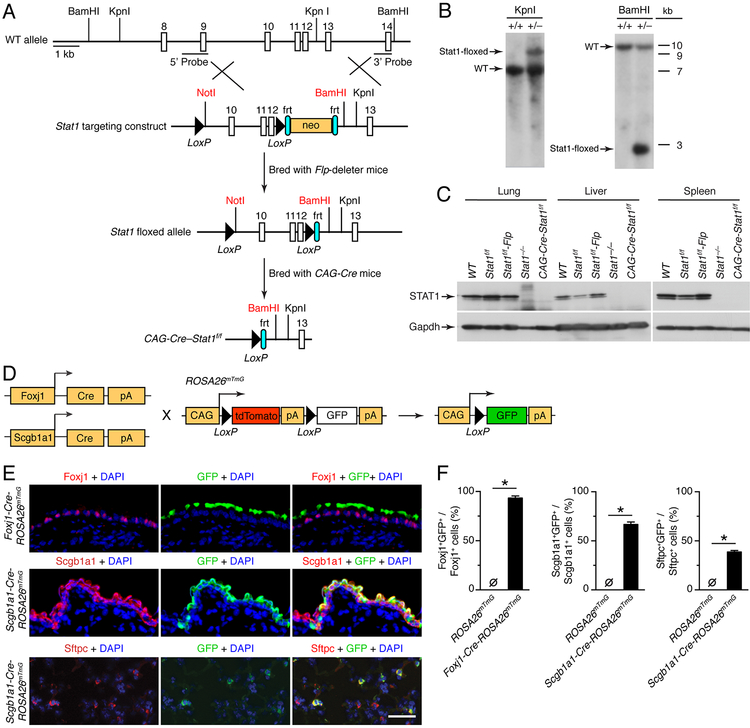
FIGURE 1.
Generation of epithelial-cell STAT1-deficient Foxj1-Scgb1a1-Cre-Stat1f/f mice. (A) Schematic of mouse wild-type (WT) Stat1 gene, Stat1 gene targeting construct, Stat1-floxed allele, and final Cre-modified Stat1flox/flox (Cre-Stat1f/f) gene locus. (B) Southern blot of tail DNA from heterozygous Stat1wt/f and control WT mice after treatment with restriction enzymes KpnI (Stat1f/f allele size of 9 kb, WT allele size of 7 kb) or BamHI (Stat1f/f allele size of 3 kb, WT allele size of 11 kb). (C) Western blot of tissue homogenates from indicated mouse strains using anti-STAT1 or anti-Gapdh Ab. (D) Breeding scheme for Foxj1-Cre or Scgb1a1-Cre mice cross to CAG-ROSA26mTmG mice to generate epithelial cell-specific CAG-GFP reporter mouse strains (Foxj1-Cre-ROSA26mTmg and Scgb1a1-Cre-ROSA26mTmG). (E) Immunostaining for Foxj1, Scgb1a1, Sfptc, and GFP and counterstain with DAPI of lung sections from Foxj1-Cre-ROSA26mTmG or Scgb1a1-ROSA26mTmG. Scale bar, 100 μm. (F) Quantitation of immunostaining for conditions in (E) along with control ROSA26mTmG mice. For (B,C,E,F), values are representative of 3 separate experiments (n≥8 mice per condition in each experiment). For (F), * indicates p<0.05.