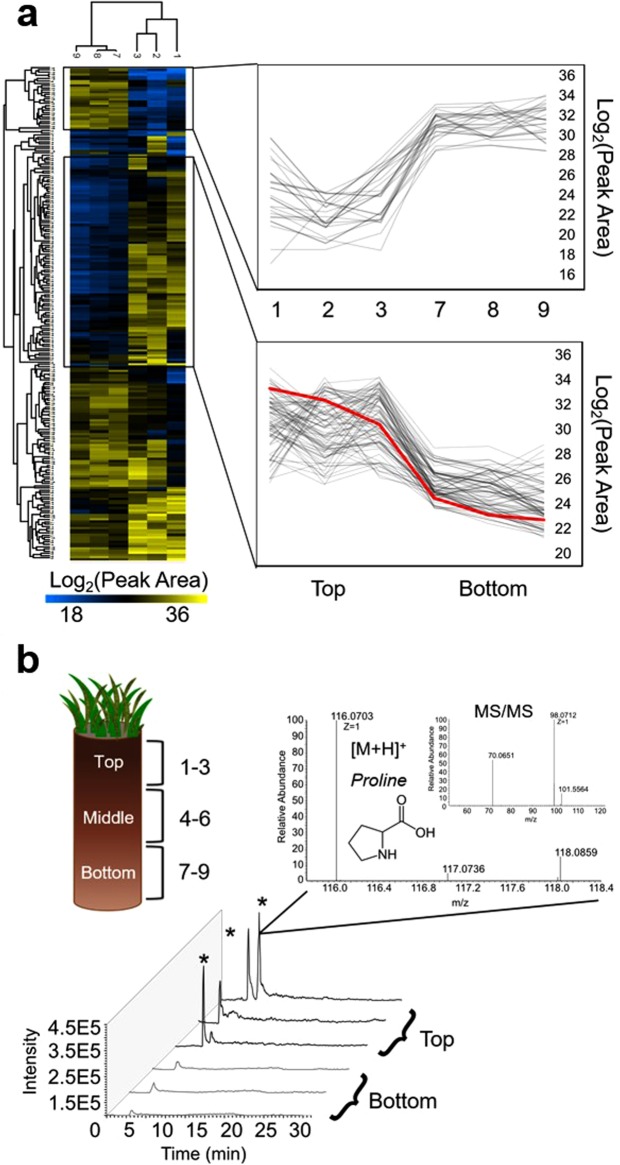
Figure 6.
(a) Heatmap of the unique IDs and normalized log2 peak areas for each abundant HQF detected by HILIC (+). The dendrogram to the left of the heatmap clusters LMW DOM metabolites that varied similarly between the top and bottom of the soil core. To the right, we call out two clusters that starkly increased or decreased with depth after normalizing to per gram dry weight soil. One feature that decreased with depth, highlighted in red, is further analyzed in (b). (b) Cross-sectional diagram of the soil core showing three depth increments and corresponding extraction replicate sample numbers. Stacked extracted ion chromatograms and mass spectra (MS1 and MS2, insets) for a feature (116.0703 m/z) detected by HILIC (+) at RT ~6.1 min. The feature was detected (intensity >1.0E5) in all six soil extracts but not in the blanks or controls. Extraction triplicates yielded similar amounts (CV < 3%) despite some peak splitting, and there was a 4-fold difference between the log2 peak areas of the top and bottom sections of the core (p-value < 0.05 by t-test). The feature was putatively identified as proline by matching the MS1 spectrum in MMCD and confirming with the MS2 spectrum in MassBank.