Figure 4.
CORE Is a Widespread Complex with Conserved Functions across Species
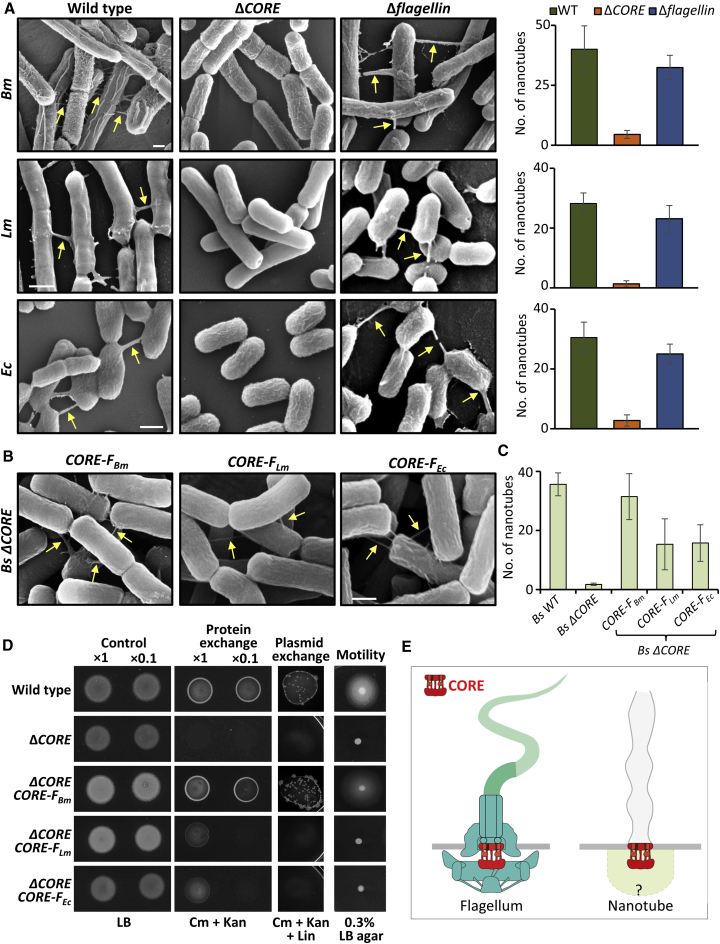
(A) Wild-type Bm (OS2), Lm (10403S), and Ec (MG1655) and their corresponding mutant strains lacking CORE (ΔCORE) or gene encoding flagellin (Δflagellin) were grown to the mid-logarithmic phase, spotted onto EM grids followed by incubation on LB agar plates for 4 h at 37°C, and visualized using XHR-SEM. Arrows indicate nanotubes. Scale bar represents 500 nm. Right: quantification of the average number of nanotubes displayed per 50 cells by the indicated strains following XHR-SEM analysis. Shown are average values and SD of at least three independent experiments (n ≥ 200 for each strain).
(B) Cells of Bs (ΔCORE) complemented with Bm flagellar CORE (CORE-FBm), Lm flagellar CORE (CORE-FLm), and Ec flagellar CORE (CORE-FEc) were processed as in (A) and visualized using XHR-SEM. Arrows indicate nanotubes. Scale bar represents 500 nm.
(C) Quantification of the average number of nanotubes displayed per 50 cells by the indicated strains following XHR-SEM analysis described in (B). Shown are average values and SD of at least three independent experiments (n ≥ 200 for each strain).
(D) Assessing molecular exchange in Bs ΔCORE strain complemented with exogenous CORE genes. For protein exchange assay, pairs of a donor (SB463: amyE::Phyper-spank-cat-spec) (CmR, SpecR) and a recipient (SB513: amyE::Phyper-spank-gfp-kan) (KanR) parental strains (wild-type) were used. The investigated strains ΔCORE and ΔCORE complemented with CORE-FBm, CORE-FLm, and CORE-FEc (controlled by Bs Pfla/che promoter) harbor the corresponding genotypes of donor and recipient. Donor and recipient strains were mixed in 1:1 ratio (at two concentrations, 1× and 0.1×) and incubated in LB supplemented with 1 mM IPTG for 4 h at 37°C with gentle shaking. Equal numbers of cells were then spotted onto LB agar (control) and LB agar containing chloramphenicol (Cm) and kanamycin (Kan) (protein exchange) and photographed after 18 h. For plasmid exchange assay, pairs of a donor (GD110: amyE::Phyper-spank-cat-spec, pHB201/cat, erm) (CmR, SpecR, MlsR) and a recipient (SB513: amyE::Phyper-spank-gfp-kan) (KanR) parental strains (wild-type) were used. The investigated strains complemented with exogenous COREs harbor the corresponding genotypes of donor and recipient strains. Cells were mixed in 1:1 ratio (concentration 1×), processed as described for protein exchange, and spotted onto LB agar containing Cm, Kan, and lincomycin (Lin) (plasmid exchange). Cells were incubated at 37°C, and colonies were photographed after 36 h of incubation. For motility assay, wild-type (PY79) and the indicated strains were grown to mid-logarithmic phase, spotted onto LB plates containing 0.3% agar, and photographed after 7 h of incubation at 37°C (motility).
(E) A schematic model depicting the modularity of CORE complexes in flagellum and nanotube. CORE-associated components of the nanotube basal body are missing. Flagellum structure was adapted from (Dietsche et al., 2016).