Figure 1.
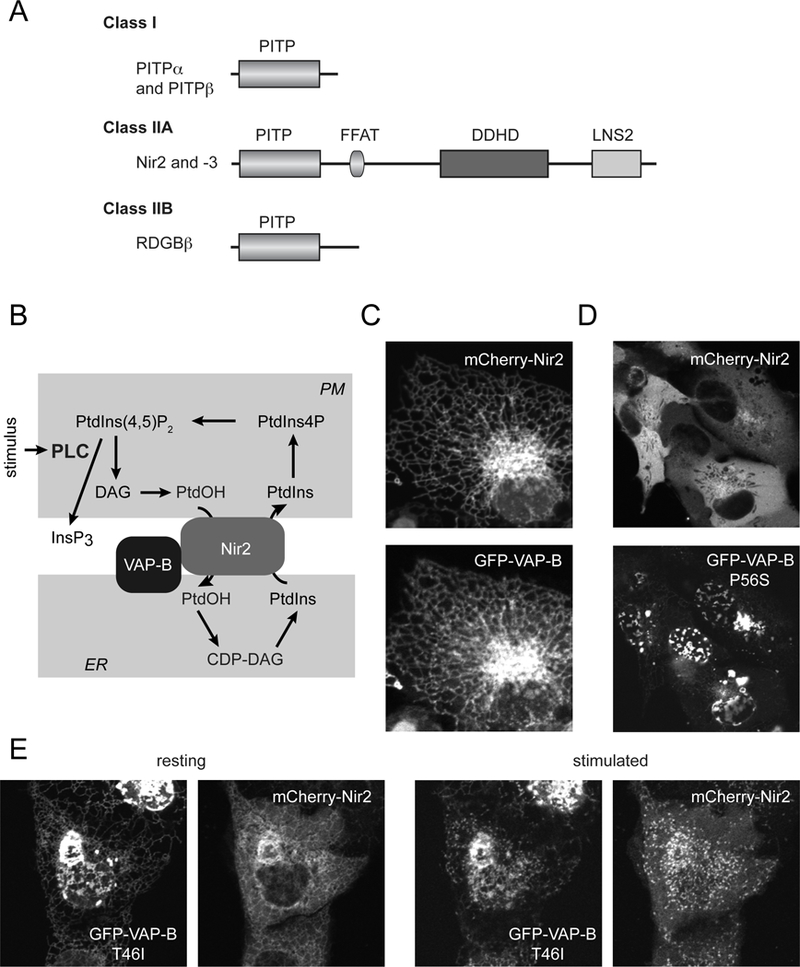
(A) Classification of PITPs. Class I PITPs comprise of PITPα and PITPβ, the latter having two splice variants. The Class IIA PITPs, Nir2 and Nir3 are homologues of the Drosophila RDGB protein (also called PITPnm1 and −2). These are larger proteins having additional domains, such as the FFAT motif (double phenylalanine in an acidic stretch), the DDHD domain and the LNS2 domain, the latter also present in lipins and binds PtdOH. Class IIB PITP is more homologous to the PITP domain of Nir2/3 than to the Class I group. (B) The proposed model of Nir2 action during PLC activation. Nir2 is recruited to the PM via its LNS2 domain (with the help of a short sequence preceding the LNS2 domain reminiscent of DAG binding sequences) and anchored to the ER via interaction via its FFAT domain with VAP proteins. Nir2 then transfers PtdIns from the ER to the PM and PtdOH in the other direction. (C) Cellular localization of Nir2 and VAP-B in resting HEK293 cells. Note the co-localization of the two proteins in the tubular ER. (D) Localization of the mutant VAP-B-P56S and Nir2 in quiescent HEK293 cells. Note the mutant VAP-B aggregates at the nuclear envelope and the lack of recruitment of Nir2 to the mutant VAP-B protein. (E) Localization of the mutant VAP-B-T46I and Nir2 in quiescent (left) or Angiotensin II-stimulated (right) HEK293 cells. Note that this mutant still attracts Nir2 to the ER even though it also shows a tendency to aggregate. After stimulation, the two proteins shows small clusters that correspond to ER-PM contact sites. The T46I mutation of the VAP-B is less disruptive than the P56S.
