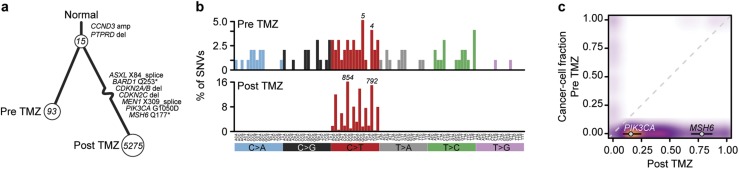
Figure 2.
(a) Sample tree showing clonal relationship of the pre-TMZ primary specimen and the posttreatment liver metastasis, with numbers in circles indicating mutations acquired from the previous branch point. The two tumors share 15 mutations. The post-TMZ sample acquired a large number of private mutations, typical of therapy-induced hypermutation. Mutations considered likely oncogenic are highlighted. (b) The substitution types (colored labels) and trinucleotide context (vertical labels) of SNVs in both samples, as a fraction of total mutations. An enrichment for C>T/G>A transitions, characteristic of alkylator-induced hypermutation, was present in the posttreatment but not pretreatment sample. Numbers above bars indicate absolute number of mutations in that bin. (c) Estimates of fraction of cancer cells harboring mutations in both samples, represented as a three-dimensional density plot, showing the large number of mutations acquired in the post-TMZ sample. Whereas the mutation in MSH6 was nearly clonal in the liver specimen, the acquired mutation in PIK3CA was found in a minority of the cell population.