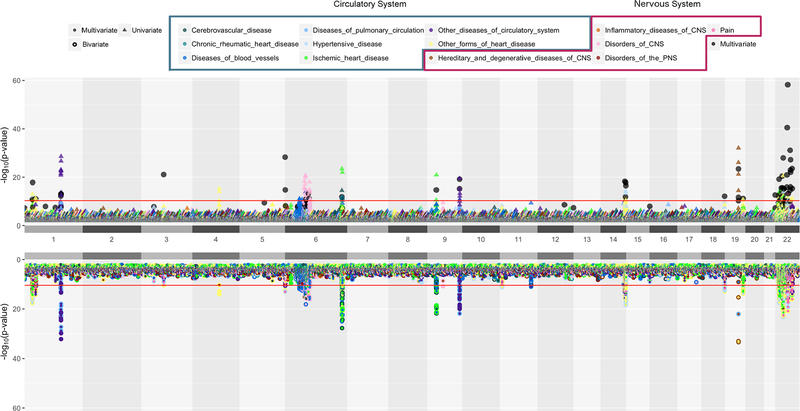
Figure 3.
Univariate, Bivariate and Multivariate Results
A position-by-position comparison of genetic associations for univariate, bivariate and multivariate methods using code modified from Hudson R package (https://github.com/anastasia-lucas/hudson). The horizontal axis represents genomic locations by chromosome and the vertical axis represents −log10(p-value). Colors represent major disease groups of circulatory and nervous systems. The top plot presents univariate results with p-value less than 0.01 in triangles and multivariate results that passed “method-specific Bonferroni” threshold in black dots. The bottom plot present bivariate analysis results in a two-colored circle, denoting the two phenotypes with which a variant is associated with. The red lines in both plots are the “family-wise Bonferroni” threshold.