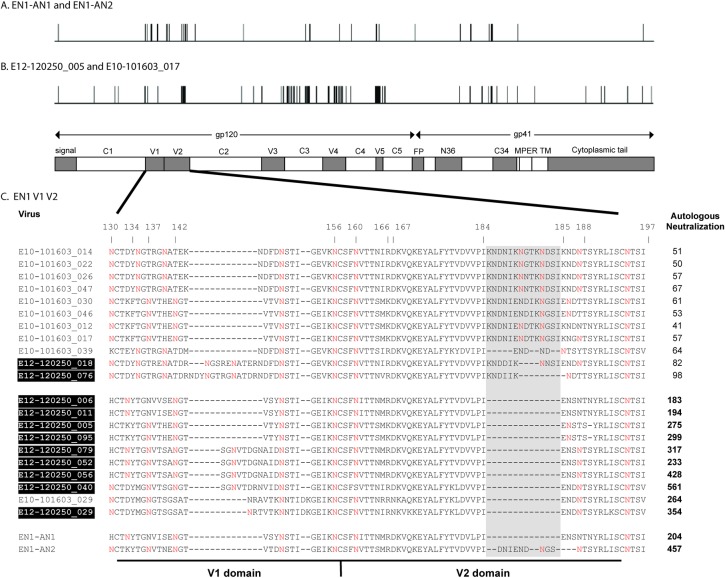
Fig 4. Characterization of the locations of sequence differences between inferred ancestral envelope proteins and authentic envelope proteins from EN1.
The location and number of sequence differences between the inferred ancestral EN1-AN1 and EN1-AN2 envelope proteins (A) and the authentic envelope proteins (E12-120250_005 and E10-101603-017) (B) are represented in horizontal diagrams of gp160 structure. EN1-AN1 and EN1-AN2 represent common ancestors of virus sequence present before and after the evolution of resistance to neutralization by autologous plasma. The E12-120250_005 and E10-101603_017 sequences are representative of authentic sequences from the most recently evolved clades of viruses sensitive and resistant to neutralization by contemporaneous autologous EN1 serum. (C) Alignment of V1/V2 sequences from EN1 highlighting V2 insertion in relation to autologous neutralization titers. Neutralization resistance in EN1 correlated with a 9–14 amino acid insertion containing two N-linked glycosylation sites between positions 184 and 185 in the V2 domain (gray shading). Shaded boxes indicate provirus sequences, unshaded labels indicate plasma virus sequences. Values in bold represent significant neutralization, as described in Table 1. EN1-AN1 and EN2-AN2 represent the inferred sequences at nodes indicated in Fig 1. Red Ns (N), indicate the locations of predicted N-linked glycosylation sites. Amino acid positions are designated with reference to the prototypic HXB2 envelope sequence.