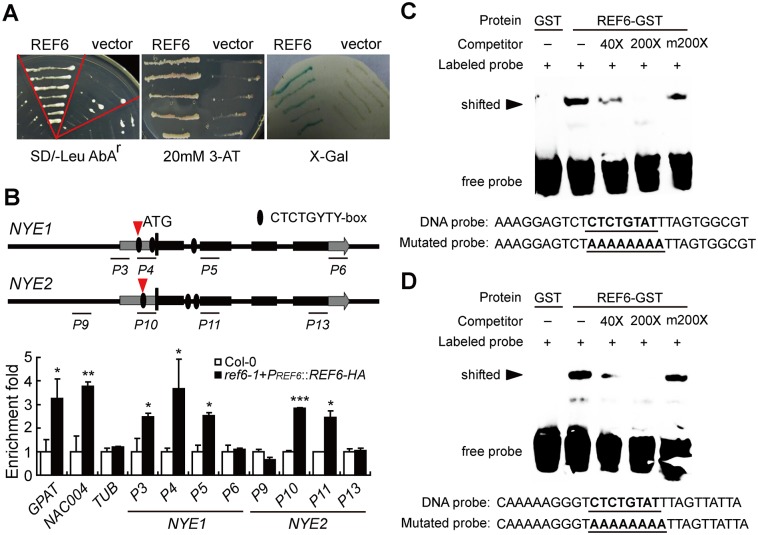
Fig 1. REF6 directly binds to both the promoters and coding regions of NYE1 and NYE2.
(A) Physical interactions of REF6 with NYE1 promoter in Y1H assays and blue-white spotting tests. (B) ChIP assays of in vivo association of REF6-HA with NYE1/2 genes in the 10-day-old seedlings of ref6-1+PREF6::REF6-HA and Col-0. Fold enrichments were calculated as the ratios of the signals in ref6-1+PREF6::REF6-HA to the signals in Col-0. Primers are listed in S4 Table. GPAT4 and NAC004 genes were used as positive controls, with TUB as a negative control. Data are mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 by paired Student’s t test. (C, D) EMSAs of in vitro binding of REF6 to the CTCTGYTY motifs within the ChIP-PCR fragments P4 and P10 of NYE1 (C) and NYE2 (D) promoters (indicated with red arrows). Probe sequences used in EMSA are shown in (C) and (D). GST-tagged REF6 was incubated with the biotin-labeled wild-type DNA probe. Competition experiments were performed by adding the excessive amounts (40× and 200×, respectively) of unlabeled DNA probe. A mutated probe was used to test binding specificity. Shifted bands, indicating the formation of DNA-protein complexes, are indicated by arrows. “-” represents absence, “+” represents presence. Sequences of both the wild-type and mutated probes are shown on the bottom of the images.