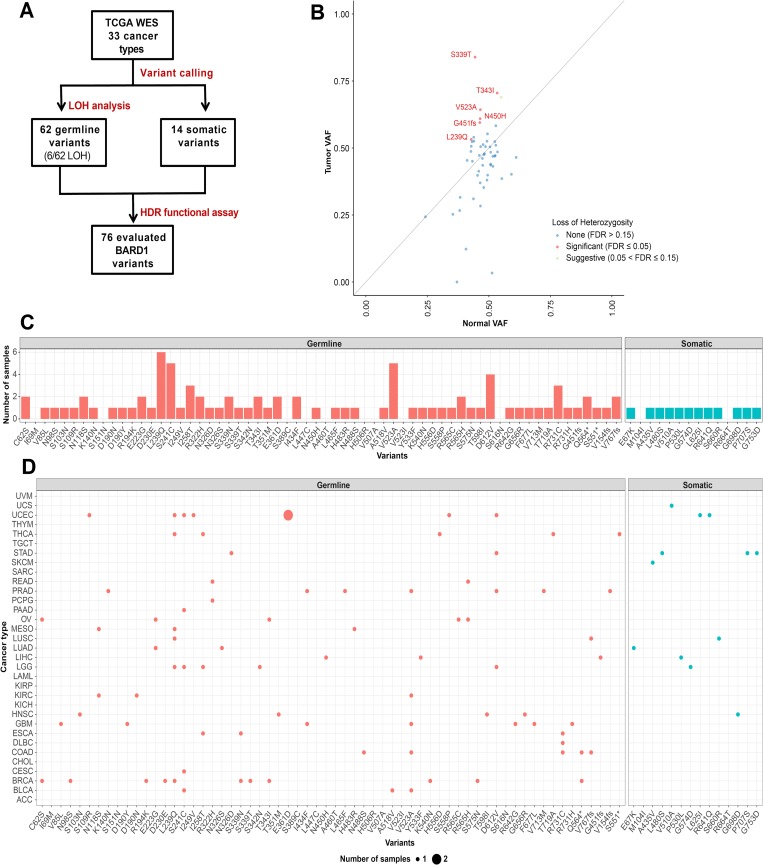
Fig 1. Selection of BARD1 missense variants for functional analysis using sequencing data of cancer patient samples.
(A) BARD1 missense variants of interest were identified in a cohort of 4,034 samples from 12 cancer types and a larger set of 10,389 TCGA samples from 33 cancer types with whole exome sequencing [19,22]. (B) Identification of LOH in BARD1 through comparison of VAF in tumor and normal samples. Each dot depicts one variant. The diagonal line denotes neutral selection of the germline variant where the normal and tumor variant allele frequencies (VAFs) are identical. LOH was considered significant at False Discovery Rate (FDR) ≤ 0.05. (C) Number of samples containing each of the 76 BARD1 variants in the 10,389 cohort [22]. (D) Number of samples affected by each BARD1 variant for each of the 33 cancer types.