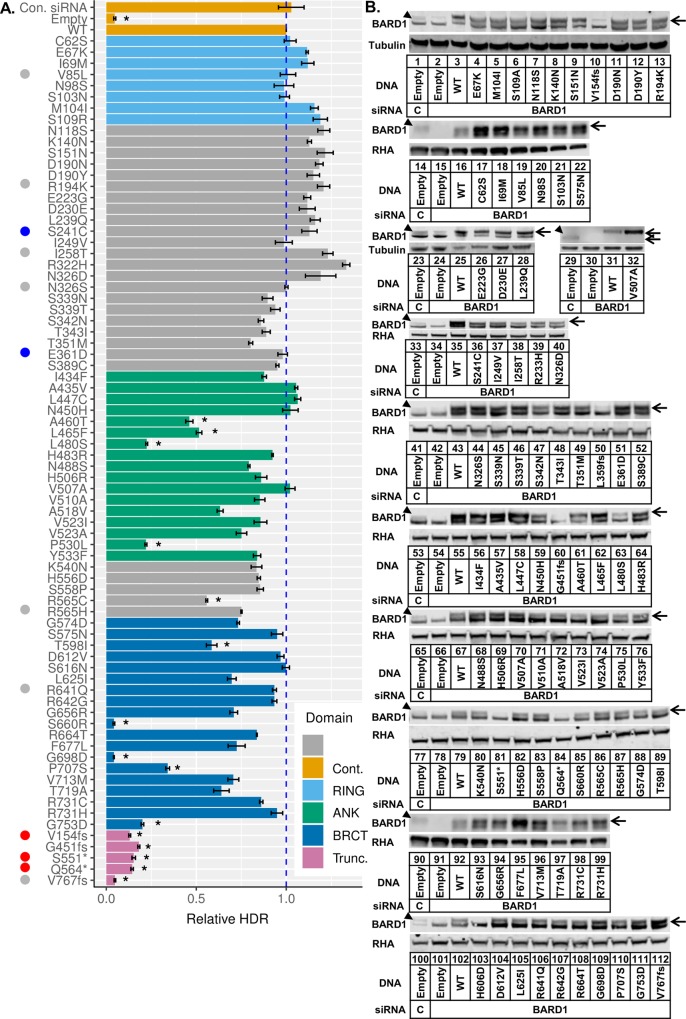
Fig 2. Functional analysis of BARD1 variants.
(A) 76 BARD1 single missense substitutions were tested for function in the HDR assay. HeLa-DR cells [16] were treated with control siRNA (lane 1) or siRNA specific to the BARD1 3'-untranslated region (UTR) (lanes 2–80) and empty vector (lanes 1,2) or BARD1 expression plasmid (lanes 3–80). Two positive controls were used: cells treated with empty vector and control siRNA (lane 1), and cells depleted of endogenous BARD1 with wild-type BARD1 rescue (lane 3). Cells treated with empty plasmid and BARD1 3'UTR siRNA were used as a negative control (lane 2). HDR function was characterized by the percentage of GFP-positive cells measured using flow cytometry. Results in each experiment (±S.E.M.) were normalized to the WT rescue (lane 3), which was set equal to 1. Results represent three independent transfections per BARD1 plasmid. Variants that are benign and pathogenic according to ClinVar are labeled blue and red respectively. Variants with conflicting interpretations are labeled gray. HDR-deficient variants are marked by an asterisk and classified by having HDR function less than 0.6 and p < 0.01 when compared with endogenous BARD1 (control siRNA) using the Student’s t-test. (B) BARD1 variants tested in the HDR assay were examined for their expression relative to endogenous BARD1. Replicates were pooled together to examine BARD1 expression. The BARD1 protein is indicated with an arrow, as the BARD1-specific band migrated more slowly than a cross-contaminating band. The endogenously expressed BARD1 in control transfections (lanes 1, 14, 23, 29, 33, 41, 53, 65, 77, 90, 100) and the depleted BARD1 without rescue (lanes 2, 15, 24, 30, 34, 42, 54, 66, 78, 91, 101) can be compared with the expression of variant BARD1 proteins as indicated. A tagged BARD1 V507A plasmid was used to confirm BARD1 expression (lanes 31 and 32, upper arrow) and migrated more slowly than endogenous BARD1 (lane 29, lower arrow). All missense variants had expression higher than the endogenously expressed BARD1.