Summary
Niche cell enwrapment of stem cells and their differentiating progeny is common and provides a specialized signaling and protective environment. Elucidating the mechanisms underlying enwrapment behavior has important basic and clinical significance in not only understanding how niches are formed and maintained, but also how they can be engineered and how they are misregulated in human pathologies such as cancer. Previous work in C. elegans found that when germ cells, which are enwrapped by somatic gonadal niche cells, are freed into the body cavity, they embed into other tissues. We investigated this phenomenon using live cell imaging and discovered that ectopic germ cells preferentially induce body wall muscle to extend cellular processes that enwrap the germ cells, the extent of which was strikingly similar to the distal tip cell (DTC)-germ stem cell niche. Enwrapment was specific for escaped germ cells, and genetic analysis revealed it did not depend on pathways that control cell death and engulfment or muscle arm extension. Instead, using a large-scale RNAi screen and GFP knock-in strains, we discovered that the enwrapping behavior of muscle relied upon the same suite of cell-cell adhesion molecules that functioned in the endogenous niche: the C. elegans E-cadherin HMR-1, its intracellular associates α-catenin (HMP-1) and β-catenin (HMP-2), and the L1CAM protein SAX-7. This ectopic niche-like behavior resembles the seed and soil model of cancer metastasis and offers a new model to understand factors regulating ectopic niche formation.
Keywords: cellular enwrapment, ectopic niche, stem cell, germ cell, E-cadherin, L1CAM, C. elegans
Graphical Abstract
Gordon et al. show that when C. elegans germ cells are made to escape from the gonad, they become selectively enwrapped by dynamic muscle protrusions. Enwrapping muscle resembles the endogenous germ stem cell niche, both morphologically and in the localization of adhesion complex members, yielding insight into the formation of ectopic niches.
Introduction
Cell and tissue boundaries are fundamental organizing mechanisms of multicellular life, but these boundaries must sometimes be breached during development and homeostasis. For example, leukocytes extravasate into tissues during wound healing and immune surveillance, macrophages and neutrophils encircle and phagocytose dying cells during apoptotic engulfment, and numerous cells fuse together to form organs [1–3]. Cell-in-cell incorporation, cell invasion, and cell fusion are not only important for normal processes, but are also misregulated in numerous pathologies, including immune disorders, developmental defects, and cancer [1,3–5]. These dynamic, intrusive cellular behaviors are often rare and difficult to visualize in their native settings [4–6]. As a result, the full repertoire of ways in which cells encapsulate or enter other cells and tissues may not yet be identified in both normal and disease contexts.
One area of active inquiry is how stem cells become embedded in their niche. Cells that are enwrapped by supportive niche cells include mouse spermatogonial stem cells and their differentiating progeny [7]; Drosophila intestinal progenitor cells [8], primordial germ cells, germ stem cells, and their progeny [9]; zebrafish hematopoietic stem and progenitor cells [10]; and C. elegans germ cell progenitors [11], stem cells [12–14], and their progeny [12]. Niche enwrapment may increase surface area to either amplify or spatially restrict niche signaling, to physically anchor cells in the niche, and to create physiologically buffered microenvironments that nurture and protect stem cells [10,13–15]. Understanding the mechanisms that regulate the establishment of physiological and ectopic niches has important implications for fertility, renewal of aged and damaged tissues, and the generation of tumor cell niches that sustain metastasis. It is unclear whether cells are capable of inducing their own enwrapment in ectopic locations, and the molecular mechanisms regulating niche cell enwrapment are poorly understood [9,11,14]. A system in which cells are endogenously enwrapped and become ectopically enwrapped under certain conditions is necessary to study these important questions.
In C. elegans mutant backgrounds that directly disrupt the gonadal basement membrane [16–19] or cause germ cell hyper-proliferation [20], germ cells escape the somatic gonad and enter the body cavity. Over time, they appear to enter other tissues [17,19], though exactly which tissues and how they enter is not known. We ruled out several known modes of tissue entry and tested a new hypothesis that the tissue-disruptive behavior of germ cells may be related to their ability to be enwrapped by their stem cell niche. By performing live-cell imaging in combination with genetic analysis to examine escaped germ cell interactions with body wall muscles, we discovered that ectopic germ cells induced cellular enwrapment by muscle cells, which resembled germ stem cell niche enwrapment. Through a large-scale RNAi screen and genome editing, we demonstrated that enwrapment by both the normal niche and by muscle were mediated by the homophilic cell adhesion receptor HMR-1/E-cadherin complex in cooperation with the SAX-7/L1CAM adhesion receptor. We found that displaced germ cells induced niche-like cellular enwrapment ectopically, thus revealing a new mechanism of inappropriate cell entry into tissues that could possibly establish de novo niches.
Results
Muscle cells dynamically enwrap escaped germ cells
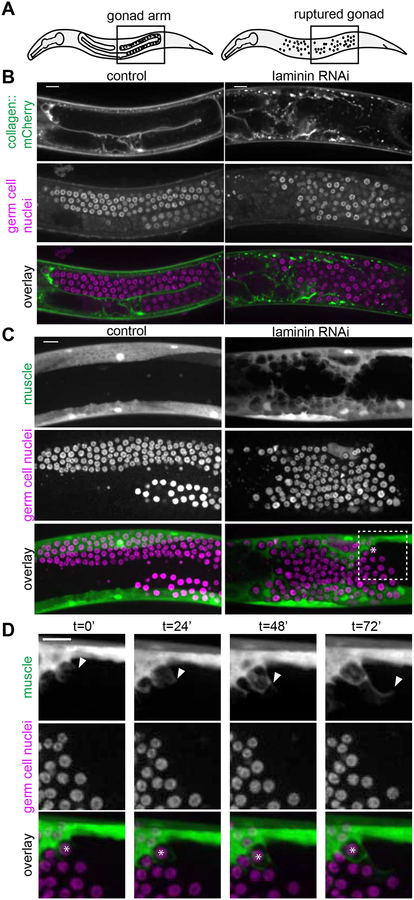
In various C. elegans mutants, escaped germ cells have been postulated to be actively invasive, to fuse with other tissues, or to be engulfed (as with apoptosis) by neighboring tissues [17–20]. We developed a robust assay to examine ectopic germ cells using RNAi targeting the epi-1/laminin α subunit beginning at the L1 larval stage, which ruptured the distal gonadal basement membrane in later larval stages (Figure S1). Loss of basement membrane released germ cells into the body cavity, which we then imaged in vivo (Figure 1A, 1B, Figure S1). We found escaped germ cells (visualized by nuclear histone marker mex-5p::H2B::mCherry) consistently embedded within body wall muscle cells by the L4 stage (n = 6/6 egl-13p::GFP muscle marked animals, Figure 1C, and n = 33/33 myo-3p::GFP::CAAX muscle marked animals with large basement membrane ruptures, Figure S3).
Figure 1: Muscle cells dynamically enwrap escaped germ cells.
(A) Schematics of L4 larval stage worms with areas of microscopy boxed. Anterior left, dorsal top. (B) Single confocal z-slices through center of gonad regions of L4 stage control (left) and animal treated with RNAi against laminin a subunit encoded by epi-1 (right). Collagen::mCherry (emb-9p::emb-9::mCherry) localizes to the basement membrane (top). Germ cell nuclei (pie-1p::GFP::H2B) typically enclosed in the gonad are freed by gonadal basement membrane rupture caused by epi-1 RNAi (middle). Overlay (bottom). Gonad ruptures occur by L4 stage with high frequency (Figure S1), and after different genetic manipulations (Figure S3, same control and epi-1 images shown as in Figure 1B). (C) Maximum intensity z-projections through 7 μm of control (left) and epi-1 RNAi (right). Muscle cells (top, egl-13p::GFP) that normally do not interact with germ cells (middle, mex-5p::H2B::mCherry) enwrap escaped germ cells after epi-1 RNAi treatment. Box and asterisk in overlay (bottom) show enlargement in Figure 1D t = 0’. (D) Maximum intensity z-projections through 7 μm stills every 24 minutes of a time-lapse experiment. Muscle (top, egl-13p::GFP) grows to enwrap germ cells (middle, mex-5p::H2B::mCherry) in an animal treated with epi-1 RNAi. Arrowhead shows progression of muscle extension. Asterisk in overlay (bottom) marks one germ cell as it becomes enwrapped by muscle. Similar time-lapse imaging in a strain with a germ cell membrane marker shows that germ cell membranes are not protrusive (Video S1 and Figure S2), and Video S2 and Video S3 show laminin RNAi and control animals as in Figure 1C. Scale bars are 10 μm.
We first asked if ectopic germ cells fused with muscle cells or were actively invasive. We conducted time-lapse imaging of muscle and germ cell membranes (myo-3p::mCherry::PLCδPH and mex-5p::GFP::PLCδPH::nos-2 3’UTR, respectively). As evidenced by the membrane-membrane contact that persists between the muscle and enwrapped germ cells (Figure S2, n = 9/9 animals maintained distinct cell membranes), we concluded that germ cells neither fused with muscle cell membranes nor entered the cytoplasm of any muscle cell, and thus were not undergoing entosis (cell-in-cell invasion) [21]. Further, the germ cells were not protrusive, nor did they actively invade between muscle cells (Figure S2; n = 0/9 animals had invasive germ cells, see Methods). Instead, we observed that muscle cells extended protrusions that enwrapped escaped germ cells in all time-lapses (Figure S2; n = 9/9 animals had protrusive muscle, Video S1). C. elegans germ cells exist in a syncytium, and it is not clear whether muscle-enwrapped cells pinch off from this syncytium or maintain their small cytoplasmic bridges ([22]) to the rest of the germ cells.
We observed the same protrusive muscle phenotype more clearly in a strain in which escaped germ cells were marked with the nuclear marker (mex-5p::H2B::mCherry; Figure 1D; n = 6/6 time-lapsed animals with protrusive muscles; compare Video S2 to control Video S3). To determine if this behavior was specifically caused by laminin RNAi, we examined ectopic germ cells with gonadal basement membrane breaks induced in other ways. Genetic loss of the basement membrane receptor dgn-1/dystroglycan causes germ cells to escape [18], and upon RNAi knockdown of dgn-1 we observed ectopic enwrapment by muscle (n = 17/17 animals with ruptures showed ectopic germ cell enwrapment, Figure S3). Germ cell hyperproliferation also causes gonadal rupture, and gonad ruptures in gld-2(q497) gld-1(q485) double mutants [20,23] led to ectopic enwrapment (Figure S3, n = 12/14 animals had enwrapped germ cells). Thus, the presence of escaped germ cells in the body cavity alone was sufficient to trigger enwrapping behavior by muscles. We cannot rule out that muscle basement membrane disruption inherent in the RNAi experiments, or potential mechanical damage to the muscle that results from the organ failure of the gonad in all experiments, contributes to the protrusive muscle phenotype. Still, we only observe muscle protrusions in the presence of escaped germ cells with these treatments, and not along their entire length. Regardless, we definitively establish that it is the muscle, and not the germ cells, that acquires an abnormal behavior when the two interact.
Muscle cell enwrapment is specific for ectopic germ cells
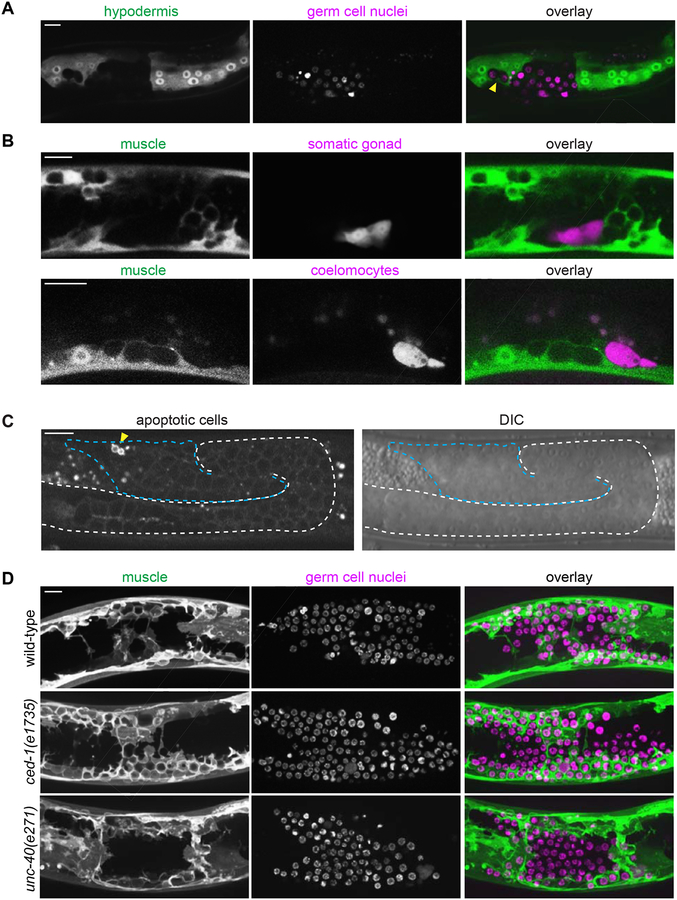
Previous transmission electron microscopy studies showed that escaped germ cells enter muscle and gut tissue [17–19]. We performed live-cell imaging with markers for muscle (Figure 1), and two other major tissues—the gut (ges-1p::GFP) and hypodermis (mlt-10p::GFP-PEST). We did not observe ectopic germ cells enwrapped by gut cells (n = 0/11 animals; Figure S4). However, germ cells were occasionally enwrapped by the hypodermis (n = 5/38 animals; Figure 2A). In these cases, fewer than ten germ cells were observed enwrapped by the hypodermis in each animal. In contrast, muscle enwrapped an average of 44 germ cells per animal (n = 13 animals). Together, these observations indicate that the muscle cells most readily enwrap escaped germ cells.
Figure 2: Ectopic enwrapment is a specific response to germ cells and is distinct from engulfment and muscle arm extension.
(A) In epi-1 RNAi-treated animals, the hypodermis (hyp7) (mlt-10p::GFP-PEST) occasionally enwraps germ cells (arrowhead, mex-5p::H2B::mCherry), but intestine does not (Figure S4). A 10 μm maximum intensity z-projection is shown. Scale bar is 10 μm. (B) In epi-1 RNAi-treated animals, muscle cells (myo-3p::mCherry) did not enwrap dorsal uterine cells (top, marked by zmp-5p::GFP) or coelomocytes (bottom, marked by unc-122p::GFP). In (B) and (C), germ cells were identified by DIC (see Methods). Single confocal z-slices shown. Scale bars are 10 μm. (C) A marker for engulfment of apoptotic cells (hsp-16.2p::TTR-52::mCherry) was expressed by only a small fraction (arrowhead) of escaped germ cells (blue dashed line) following heat shock. A single confocal fluorescent slice and corresponding DIC image are shown. A white dashed line indicates the intact gonadal basement membrane evident by DIC; the break in the dashed line indicates a break in the gonadal basement membrane. Scale bar is 15 μm. (D) There was no difference in enwrapment between control animals (top), and mutants for the engulfment pathway (middle) or muscle-arm extension pathway (bottom) treated with epi-1 RNAi. Muscle cells (left, myo-3p::GFP::CAAX) enwrapped escaped germ cells (center, mex-5p::mCherry::H2B) in the ced-1/Draper engulfment-defective mutant ced-1(e1735) and in the muscle arm extension-defective mutant unc-40(e271). Maximum intensity z-projections of the superficial-most 5 μm containing mCherry-labeled germ cells are shown for muscle and germ cell fluorescence signals and as an overlay (right). Scale bar is 10 μm.
We next examined whether other cells in the body cavity induce this enwrapment by muscles. We found that body wall muscles did not enwrap coelomocytes (marked by unc-122p::GFP; n = 0/15 animals, including n = 0/2 coelomocytes adjacent to enwrapped germ cells; Figure 2B), which are phagocytic cells that are thought to function similarly to vertebrate macrophages [24]. In addition, ectopic somatic gonad cells (zmp-5p::GFP) freed by gonad rupture were never enwrapped (n = 0/19 animals including n = 0/9 somatic gonad cells adjacent to enwrapped germ cells, Figure 2B). These data suggest that ectopic enwrapment is specific for escaped germ cells.
Ectopic germ cell enwrapment is not directed by engulfment or muscle arm extension pathways
We next tested if ectopic germ cells stimulate enwrapment through two known pathways by which cells are induced to extend cellular protrusions towards other cells: engulfment of cells undergoing apoptosis [25] and muscle arm extension by which muscles contact motor axons to form the neuromuscular junction [26]. We found that most ectopic germ cells do not express the TTR-52::mCherry marker of apoptosis [25] (n = 10 animals, total of 13/604 escaped germ cells positive for TTR-52::mCherry; Figure 2C). Further, worms with the null allele for the engulfment receptor ced-1/Draper ced-1(e1735) [27] all exhibited robust enwrapment of germ cells (n = 22/22 mutant animals with enwrapment, compared to 23/23 controls, Fisher’s exact test, two tailed p = 1.000, Figure 2D). We thus rejected the hypothesis that muscle enwrapment of germ cells is phagocytosis of dying cells using the apoptotic engulfment pathway. We next examined the function of the netrin receptor UNC-40/DCC, which mediates muscle arm extension to motor axons [26], as this is a developmentally regulated protrusive muscle behavior that could potentially be co-opted to mediate ectopic muscle-germ cell contact. Animals with the null allele for unc-40(e271) had extensive germ cell enwrapment (n = 22/27 mutant animals showed full enwrapment of escaped germ cells by dorsal body wall muscle compared to n = 10/10 controls, Fisher’s exact test, two tailed p = 0.2954, Figure 2). We concluded that ectopic germ cell enwrapment is neither mediated by mechanisms controlling muscle arm extension nor apoptotic engulfment.
Ectopic germ cell enwrapment resembles niche enwrapment, but does not support germ cell proliferation
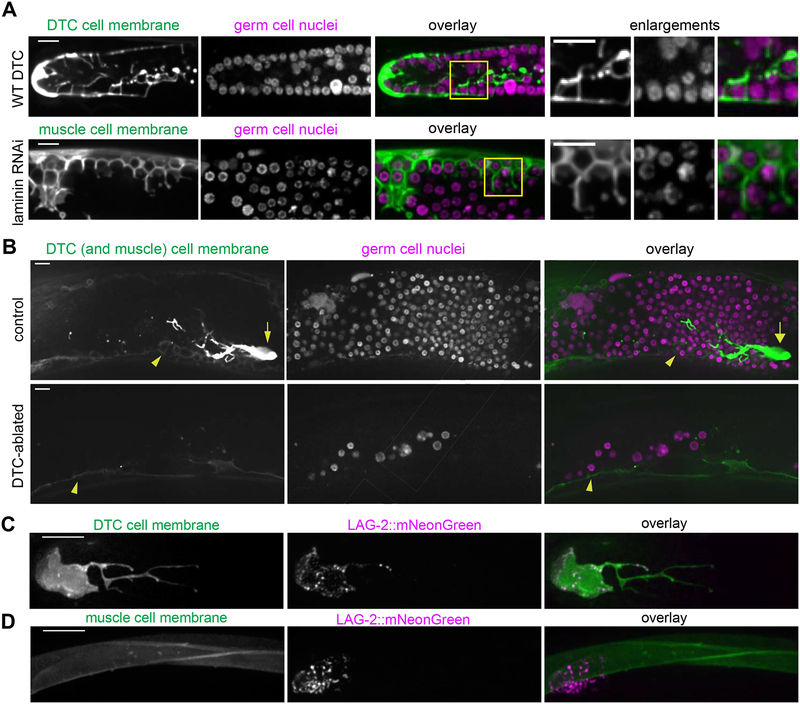
C. elegans primordial germ cells, germ stem cells, and differentiating progeny are enwrapped by somatic gonadal support cells [11–14]. The distal tip cell (DTC) enwraps the germ stem cells to form the germ stem cell niche [13,14], and its many short protrusions (called the DTC plexus, [13]) resemble ectopic muscle enwrapment of germ cells (Figure 3A). We hypothesized that the ectopic enwrapment of germ cells by muscle might recapitulate aspects of germ cell-niche interactions and investigated whether the muscle can serve as an ectopic stem cell niche. Active signaling through the Notch pathway receptor GLP-1 is necessary and sufficient to maintain germ cells in a stem-like undifferentiated state [28]. The Notch ligand LAG-2, expressed by the DTC, is required for germline expansion [29–31]; the Notch ligand APX-1 is also expressed by the DTC [31] but its endogenous expression is not sufficient to rescue lag-2 loss of function [31]. Enwrapment by the DTC facilitates activation of Notch targets [14]. A lag-2p::GFP::CAAX transgene drove low levels of GFP expression in body wall muscle (Figure 3B, arrowheads). To test whether the muscle provided a niche-like cue leading to proliferation of the ectopic germ cells, or alternatively if proliferation instead remained dependent on the normal DTC niche, we ablated the DTCs in L3/L4 larvae treated with RNAi to cause gonad ruptures. We asked whether the enwrapping muscle cells could support germ cell proliferation in the absence of the DTC. The germ cell population was dramatically diminished (n = 4/4 animals with few germ cells, average of 32.75±6.35 germ cells in the center of the ruptured gonad arm) over the 48 hours following DTC ablation relative to controls (n = 4/4 animals with many germ cells, average of 291.50±18.60 germ cells in the center of the ruptured gonad arm, see Methods and Figure 3B). We concluded that enwrapment by the muscle was not sufficient to sustain ectopic germ cells and that their survival and proliferation required the endogenous signal from the DTC. We noted that the DTC, which normally has long, branching projections extending proximally in an intact gonad [13,14] (Figure 3A), sends projections in all directions when the gonad ruptures (Figure 3B and Figure S5).
Figure 3: Ectopic germ cell en wrapment resembles niche enwrapment but does not support germ cell proliferation.
(A) Maximum intensity z-projections through 5 μm of the DTC central plexus (top left, lag-2p::GFP::CAAX) and body wall muscle cell membrane (bottom left, myo-3p::GFP:: CAAX). Germ cells (second column) are enwrapped both in the endogenous niche (top) and ectopically by muscle (bottom). Boxes in overlay (third) show areas of enlargement (right) in which both DTC (top) and muscle (bottom) have thin, enwrapping projections that surround germ cells. (B) Control (top) animal expressing lag-2p::GFP::CAAX (left) in the DTC (arrow) and muscle (arrowhead) after epi-1 RNAi have a robust germ cell population (center, mex-5p::H2B::mCherry). DTC-ablated animals (bottom) had few germ cells 48 h post-ablation compared to controls. Single confocal z-slices are shown. Full projection of the control animal also appears in Figure S5. (C) The DTC niche (left, lag-2p::myr::tdTomato) expresses the CRISPR/Cas9-tagged Notch ligand LAG-2::mNeonGreen (center), which localizes in a punctate pattern in the DTC membrane in one-day adults. Maximum intensity z-projections through 2.5 μm of confocal slices are shown. (D) Body wall muscles (left, myo-3p::GFP::CAAX) do not express LAG-2::mNeonGreen (center, punctate DTC expression visible), overlay (right). Maximum intensity z-projections through 5 μm of confocal slices are shown. Scale bars are 10 μm.
The muscle did not promote the proliferation of ectopic germ cells in the absence of the DTC, yet it expressed lag-2p::GFP::CAAX from an integrated array reporter (Figure 3B, arrowheads), suggesting lag-2 may be expressed by muscle. This transcriptional reporter, however, may not fully recapitulate endogenous lag-2 expression. To examine endogenous expression, we tagged the lag-2 gene with mNeonGreen using CRISPR/Cas9-mediated homologous recombination [32,33]. The homozygous strain was viable and fertile, while lag-2 loss of function mutants arrest and die as L1 larvae [34], assuring us that the edited allele encodes a functional LAG-2. LAG-2::mNeonGreen decorated the germ cell-facing membrane of the DTC, including much of the membrane that makes up its long, enwrapping processes (n = 9/9 animals, Figure 3C). However, LAG-2::mNeonGreen was not detected in the muscle (n = 13/13 animals, Figure 3D), so the lag-2p::GFP::CAAX expression we observed in Figure 2 likely resulted from aberrant overexpression of the transgene. Together, these observations indicated that while the muscle cells display niche-like enwrapping behavior, they neither express the Notch ligand LAG-2 nor support germ cell proliferation.
DTC adhesion to germ cells is mediated by E-cadherin and L1CAM
A common feature of niches in mice, zebrafish, and Drosophila is the cellular enwrapment of stem cells and their progeny by support cells [7,8,10,15,35,36]. We hypothesized that the cell surface proteins that control enwrapping behavior were commonly expressed in both the niche and muscle, thereby priming the muscle to respond to the presence of germ cells. We sought the direct mediators of cell-cell interactions between the somatic and enwrapped germ cells.
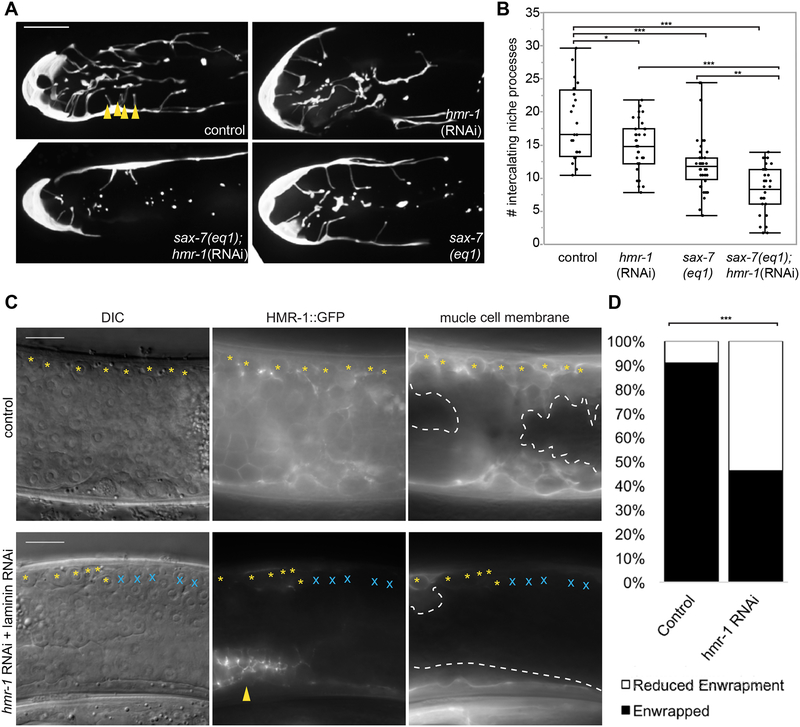
To identify genes that directly mediate DTC-germ cell enwrapment, we undertook a DTC-specific large-scale visual RNAi screen targeting 2,289 genes. Loss of 35 genes led to a qualitatively dramatic reduction in DTC length in at least 9/10 treated animals (see Methods and Table S1). To identify genes that might also mediate ectopic enwrapment by muscle, we cross-referenced these 35 genes with a body wall muscle transcriptome [37] and found six candidates (Table S1, unshaded rows), including hmr-1, which encodes the C. elegans ortholog of the cell adhesion molecule E-cadherin [38]. Drosophila E-cadherin mediates embryonic germ progenitor cell enwrapment by support cells and is one of the only genes known to mediate a niche-like enwrapping behavior [9,39]. Because null mutations in hmr-1 cause embryonic lethality [38], we examined young adult worms that were treated with hmr-1 RNAi beginning at the L1 stage. Knockdown of hmr-1 (Figure 4A) resulted in a >20% reduction in average DTC enwrapment in one day adults as quantified by the number of short intercalating processes that extend among the distal germ cells (Figure 4B). This phenotype is a more direct measure of enwrapment than total DTC length, though it cannot be measured in a high-throughput screen. As hmr-1/E-cadherin and the cell adhesion molecule sax-7/L1CAM function synergistically during C. elegans gastrulation [40], we next examined whether loss of sax-7 also resulted in enwrapment defects. Animals with the null mutation sax-7(eq1) (Figure 4A) had a ~40% reduction in the average number of short intercalating processes, and hmr-1/cadherin RNAi knockdown in the null sax-7(eq1) background resulted in a >50% reduction in the average number of short intercalating processes relative to wild type (Figure 4B). Together, these results indicated that HMR-1/E-cadherin and SAX-7/L1 CAM adhesion systems function together to promote germ cell enwrapment by the niche.
Figure 4: HMR-1/E-cadherin mediates ectopic enwrapment of germ cells by muscle.
(A) A control DTC (upper left, L4440 vector) has more intercalating processes than the DTC of an animal with hmr-1(RNAi) in the sax-7(eq1) background (lower left), compared to mild defects observed in sax-7(eq1) animals (lower right) and hmr-1(RNAi) alone (upper right). Intercalating processes (arrowheads) are quantified in (B). Maximum intensity core z-projections through 10 μm of confocal slices are shown. (B) Box plots quantifying intercalating processes for genotypes in (A). *p < 0.05, **p < 0.005, ***p < 0.0005, n ≥ 24 animals for each group, ANOVA followed by Tukey-Kramer HSD test. (C) Animals treated with epi-1 RNAi to induce rupture (left, ectopic germ cells visible in DIC image) expressing CRISPR/Cas9-tagged HMR-1::GFP (center) and muscle membrane marker (right, myo-3p::mCherry::PLCδPH). Control (top) shows full enwrapment of all germ cells along the dorsal body wall (yellow asterisks). RNAi against hmr-1 (bottom) before epi-1 RNAi causes reduced HMR-1::GFP (center, except in uterus, arrowhead), and reduced enwrapment of germ cells (germ cells that are not enwrapped marked by blue X). RNAi treated animals also lack body-crossing muscle (white dashed line) protrusions. RNAi against hmr-1 not only decreased the number of ectopic germ cells enwrapped, but also the amount of muscle cell membrane forming muscle protrusions, which explains why fluorescence in the right RNAi panel is dimmer than the control. (D) The number of animals with full enwrapment is decreased after hmr-1 RNAi treatment. Fisher’s exact text, two-tailed ***p < 0.0005. Wide field DIC and fluorescent images are shown. The candidate gene hmr-1 was identified with an RNAi screen, see Table S1. Scale bars are 10 μm.
HMR-1/E-cadherin mediates ectopic enwrapment of germ cells by muscle
We next investigated whether E-cadherin also regulates ectopic enwrapment. We developed a two-generation sequential double RNAi method to reduce hmr-1 function in the context of ectopic enwrapment. This experiment was performed on a strain carrying the muscle membrane marker myo-3p::mCherry::PLCδPH and a CRISPR/Cas9-tagged HMR-1::GFP allele [41] to assess RNAi knockdown (see Methods). Animals with visible dorsal gonad ruptures (Figure 4C, left) and knockdown of hmr-1 confirmed by loss of HMR-1::GFP signal (Figure 4C, bottom center) were imaged and scored for the amount of enwrapment of ectopic germ cells on the dorsal side. Among control animals not treated with hmr-1 RNAi, 30/33 animals showed full enwrapment of all of the escaped germ cells bordering the dorsal body wall muscles; the remaining three showed partial enwrapment. Among experimental animals treated with hmr-1 RNAi with knockdown confirmed by complete or near-complete loss of HMR-1::GFP in the muscle and germ cells, only 12/26 animals showed full enwrapment of dorsal escaped germ cells (Figure 4D, statistically different from control, Fisher’s exact test, two-tailed p = 0.0003). Furthermore, 4/26 RNAi-treated animals with the most robust knockdown of HMR-1::GFP lacked body-crossing muscle protrusions (Figure 4C, compare dashed lines). This failure of dramatic muscle remodeling by ectopic germ cells was not observed under any other conditions in which the gonadal basement membrane rupture was large enough to free many germ cells (for example, ced-1 and unc-40 mutants in Figure 2D). We concluded that hmr-1 mediates both normal niche germ stem cell enwrapment and ectopic muscle germ cell enwrapment.
Cadherin, α- and β-catenin, and L1CAM localize to DTC and muscle protrusions contacting germ cells
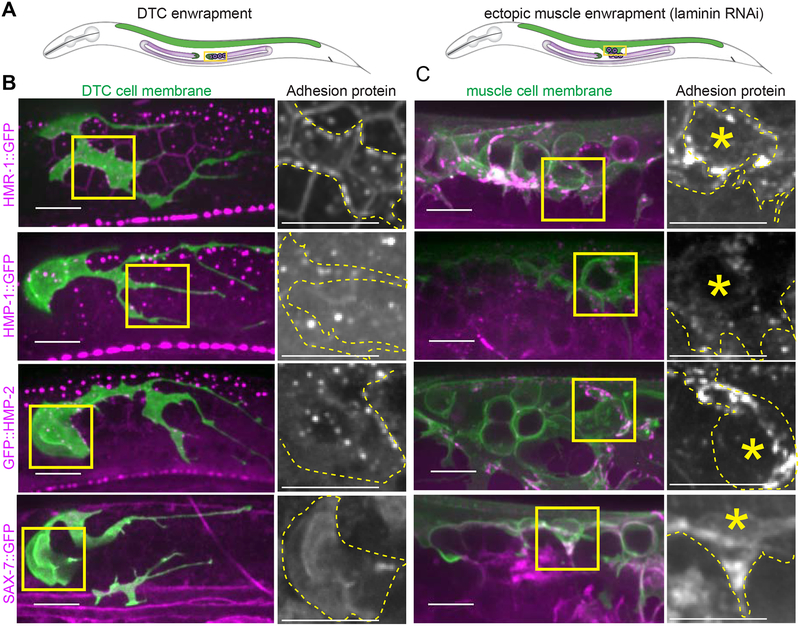
Classical cadherins are homophilic cell-cell adhesion molecules [41] and L1CAM most often mediates homophilic cell-cell adhesions as well [42]. We thus next investigated whether these proteins localized to points of contact between the DTC and germ cells (Figure 5A). A functional, endogenously GFP-tagged hmr-1 allele encoding HMR-1::GFP [41] was expressed in the DTC and germ cells and was enriched at the sites of germ cell-germ cell and DTC-germ cell contact (n = 16/16, Figure 5B, top left). Similarly, functional, endogenously tagged isoforms A-C of α-catenin (HMP-1::GFP n = 19/19 [41]) and β-catenin (GFP::HMP-2 n = 11/11 [41]), which associate with E-cadherin [41], were also expressed in the DTC and germ cells and localized to membrane contact sites (Figure 5B, left, second and third rows). HMP-1::GFP only tags half of the HMP-1 isoforms, which may make it more difficult to see. However, single confocal slices clearly show that it is present in the DTC, muscle, and germline (Figure S6). A fosmid transgene containing sax-7p::TY1::EGFP [43] was expressed in the DTC and localized to the membrane (n = 7/7, Figure 5B, bottom left). Protein expression is reported in the germline [44], but the transgene was not detected there, possibly as a result of germline transgene silencing [45]. Consistent with a role for these proteins in ectopic enwrapment, all endogenously tagged adhesion proteins localized to the muscle protrusions enwrapping escaped germ cells (Figure 5B, right, HMR-1 n = 10/10, HMP-1 n =10/10, HMP-2 n = 10/10, SAX-7 n = 11/11, Figure 5C). Further, HMR-1::GFP was enriched in the leading edges of muscle protrusions compared to the same muscle farther from the site of germ cell contact (an average of 1.6-fold brighter fluorescence at the leading edge, n = 10 animals, one tailed Wilcoxon Signed Rank test is significant at p ≤ 0.05.). Taken together, the functional requirements for hmr-1 and the localization of tagged HMR-1, HMP-1, HMP-2, and SAX-7 suggested that ectopic germ cells and enwrapping muscle exploit the same adhesion systems as those mediating germ cell enwrapment by the niche (Figure 6).
Figure 5. Cadherin, α- and β-catenin, and L1CAM localize to DTC and muscle protrusions contacting germ cells.
(A) Schematics show young adult worms with the distal intact gonad (left) and ruptured gonad region (right) boxed, adjacent to the dorsal body muscle (green). (B) lag-2p::mKate:: PLCδPH DTC membrane marker (green) coexpressed with GFP-tagged adhesion proteins (magenta). Yellow boxes show areas of enlargement. Enlarged images show adhesion protein channel only, with the DTC membrane outlined by a yellow dashed line. (C) myo-3p::mCherry:: PLCδPH muscle membrane marker (green) coexpressed with GFP-tagged adhesion proteins (magenta). Yellow boxes show areas of enlargement. Enlarged images show adhesion protein channel only, with muscle membrane outlined in yellow dashed line and focal germ cell highlighted with asterisk. All images are maximum intensity 2.5 μm z-projections. Additional HMP-1::GFP single slices shown in Figure S6. All scale bars are 10 μm.
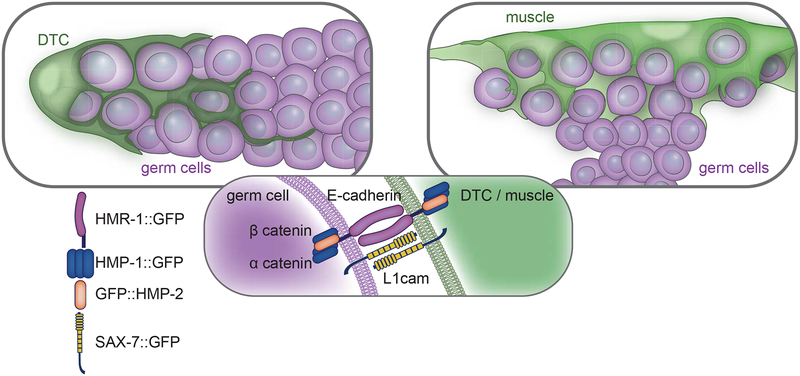
Figure 6. Endogenous and ectopic enwrapment elicited by germ cells relies on a common molecular adhesion system.
A schematic of endogenous (left) and ectopic (right) enwrapment of germ cells (magenta) by somatic cells (green). Adhesion complex components localize (center) to somatic cell and germ cell membranes.
Discussion
Niche enwrapment of stem cells and their differentiating progeny is a common, yet poorly understood, aspect of diverse niches that is thought to create a tightly regulated signaling, adhesive, and protective environment [10,11,14,15]. During zebrafish development, circulating hematopoietic stem and progenitor cells (HSPCs) trigger endothelial cell remodeling in the caudal hematopoietic tissue to form a pocket that constitutes the perivascular niche [10]. The mechanisms that direct enwrapment of HSPCs in the caudal region are not known. More is known about the molecular basis of niche enwrapment in Drosophila. In the Drosophila testes, germ cells secrete the epidermal growth (EGF) ligand Spitz, which stimulates the EGFR on somatic cyst cells and directs process extension and enwrapment through Rac GTPase activation [46]. Drosophila embryonic germ cell enwrapment is dependent on E-cadherin expressed in both soma and the germ cells [9], similar to the E-cadherin-based adhesion we observed in both niche and ectopic enwrapment of C. elegans germ cells.
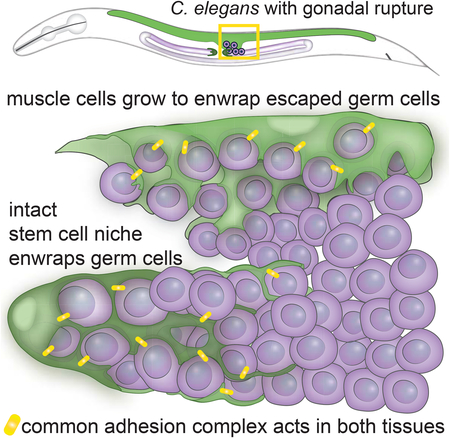
Ectopic stem cell niches have been described in Drosophila and C. elegans germlines where the normal niche is expanded by misregulation of stem cell cues, shifted in its usual position, or inappropriately occupied by other cells that respond to stem cell niche cues [47–51]. A latent niche has also been observed in the C. elegans gonad in mutant backgrounds that place undifferentiated germ cells in contact with other somatic gonad cells that express Notch ligands, which are sufficient to promote the stem-like fate in an improper location [52]. Multiple independent studies have found when C. elegans germ cells escape the gonad and enter the body cavity, they embed in other tissues [17,18,20]. By using live-cell imaging, we discovered that the germ cells induce muscle cells to extend cellular processes that intercalate among germ cells, and that this remodeled muscle cell appears similar to the normal germ stem cell niche, though the muscle does not express the crucial stemness factor LAG-2 and is therefore not a fully functional niche. Our work extends the notion of ectopic niches beyond the mispositioning of niches or stem cells within an organ to show that ectopic germ cells induce dramatic niche-like cellular enwrapment in a completely distinct tissue—the body wall muscle. These results suggest that misplaced stem and progenitor cells have the capacity to physically induce remodeling of distant tissues into niche-like microenvironments.
Ectopic enwrapment may arise from common molecular signatures between the muscle and DTC niche. We identified the homophilic adhesion molecule HMR-1/E-cadherin as a key regulator of germ cell enwrapment by both the DTC and body wall muscle. HMR-1 and its intracellular complex components HMP-1/α-catenin and HMP-2/β-catenin localize to sites of DTC-germ cell and muscle-germ cell contact, and loss of HMR-1 disrupts both normal and ectopic enwrapment. Further, we discovered that the adhesion molecule SAX-7/L1CAM also localizes to sites of cell-cell contact during normal and ectopic enwrapment and functions redundantly with HMR-1 to promote DTC-germ cell enwrapment. HMR-1 and SAX-7 function redundantly in early embryogenesis as well [40], suggesting that as an adhesion system is redeployed in different tissues, those tissues may be primed to interact if they are brought into contact by aberrant development. The C. elegans muscle might be especially receptive to enwrap ectopic germ cells, as body wall muscles form muscle arm extensions that reach motor axons [26], and thus harbor the intracellular cytoskeletal machinery for membrane extension even if ectopic enwrapment is not mediated by the muscle arm extension UNC-40/DCC signaling pathway. Additionally, the muscle endogenously expresses SAX-7, as well as HMR-1 and its intracellular complex members [37]. SAX-7 and HMR-1 seem to be necessary but not likely sufficient to induce enwrapment, as HMR-1 and SAX-7 are expressed broadly and are present in the somatic gonad cells that do not become ectopically enwrapped. Thus we expect unknown factors might be required to trigger ectopic enwrapment. It is important to note that the muscle’s ability to respond to the germ cells does not arise from muscle-cell-autonomous genetic changes, but rather from the response of the muscle to an abnormal cue from ectopic germ cells. The protrusive myoepithelial behavior is reminiscent of a mouse mammary gland cancer model in which a genetically wild-type myoepithelium actively protrudes and enwraps escaping luminal cells expressing a prometastatic gene, and requires P-cadherin to do so [53]. Similar interactions are likely important for the establishment of the metastatic niche in human cancers, in which the ability and specificity of secondary sites to support tumor growth derives both from intrinsic qualities of those sites and changes at those sites that are induced by the cancer cells themselves [54,55]. Insight into ectopic niche formation might also be exploited clinically to establish de novo niches for regeneration and tissue engineering purposes. Thus, ectopic germ cell enwrapment in C. elegans might serve as a robust model for identifying key signaling and intrinsic factors that may inform the design of synthetic niches for tissue repair, as well as therapeutic disruption of ectopic niches in diseases such as cancer.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David R. Sherwood (david.sherwood@duke.edu). Strains containing mNeonGreen can only be distributed to labs with a mNeonGreen license from Allele Biotechnology. Strains sourced from Caenorhabditis Genetics Center (CGC) are to be requested directly from CGC.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained on standard NGM media at 16, 18, 20, or 23°C and fed E.coli OP50. For RNAi experiments, wild type controls were fed E. coli HT115(DE3) containing the L4440 plasmid (see RNAi experiments, STAR Methods). All animals scored were hermaphrodites (as males have structurally different gonads) at the ages specified in the text.
Strains
In strain descriptions, we designate linkage to a promoter with a p following the gene name and designate promoter fusions and in-frame fusions with a double semicolon (::). The transgenes and alleles used in this study appear in the Key Resources Table. All strains with multiple genetic elements were generated for this study by crossing the cited strains (Source column of Key Resources Table) with genetic elements created for this study, and their Identifier is given according to the C. elegans naming convention of a two-letter lab code (D. Sherwood lab strains begin with NK) followed by a number. Some integrated strains (xxIs designation) may still contain unc-119(ed4) mutation and/or the unc-119 rescue gene in their genetic background, but these are not listed in the genetic background for the sake of concision, nor are most transgene 3’ UTR sequences. Further details available upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| RNAi feeding strain | Caenorhabditis Genetics Center (CGC) | HT115(DE3) |
| Ahringer RNAi Library | Source Bioscience | C. elegans RNAi Collection (Ahringer) |
| bacterial food: E. coli OP50 strain | CGC | OP50 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| hygromycin B from Streptomyces hygroscopicus | Millipore Sigma | CAS 31282-04-9 |
| 2,3-Butanedione monoxime (BDM) anesthetic | Millipore Sigma | CAS 57-71-6 |
| Critical Commercial Assays | ||
| Q5 site-directed mutagenesis kit | New England Biolabs | Catalog #E0554S |
| Experimental Models: Organisms/Strains | ||
| C. elegans unc-119(ed4) for transgene rescue | Paul Sternberg | PS3460 |
| axIs1677(pie-1p::GFP::H2B ::pgl-1 3′utr + unc-119(+)); qyIs46(emb-9p::emb-9::mCherry) X | WB Cat# JH2320, RRID:WB-STRAIN:JH2320 from CGC | NK2386 |
| qyIs86(egl-13p::GFP); naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | NK574 parent from [62] | NK1671 |
| qyIs448(myo-3p::mCherry::PLCδPH); xnSi1(mex-5p::GFP::PLCδPH::nos-2 3′UTR) II | FT598 parent from [63] | NK1865 |
| qyIs473(myo-3p::GFP::CAAX); gld-2(q497) gld-1(q485) /hT2 I; naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | WB Cat# JK2879, RRID:WB-STRAIN:JK2879 parent from CGC, [23] | NK2515 |
| zcIs18(ges-1p::GFP); naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | WB Cat# SJ4144, RRID:WB-STRAIN:SJ4144 parent from CGC | NK1655 |
| naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II; mgIs49(mlt-10p::GFP-PEST) IV | WB Cat# GR1438, RRID:WB-STRAIN:GR1438 from [64] | NK2516 |
| qyIs287[myo-3p::mCherry]; oxIs363(unc-122p::GFP) | WB Cat# EG5071, RRID:WB-STRAIN:EG5071 parent from CGC [56] | NK1676 |
| qyIs287[myo-3p::mCherry]; qyIs245[zmp-5p::GFP] | NK1803 parent from Kelley et al., in press | NK1690 |
| smIs119(hsp::ttr-52::mCherry) | [25] | transgene smIs119 |
| qyIs473(myo-3p::GFP::CAAX); ced-1(e1735) I; naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | WB Cat# CB3203, RRID:WB-STRAIN:CB3203 parent from CGC | NK2185 |
| qyIs473(myo-3p::GFP::CAAX); unc-40(e271) I; naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | WB Cat# CB271, RRID:WB-STRAIN:CB271 parent from CGC | NK2037 |
| qyIs353(lag-2p::GFP::CAAX); naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | nasi2 transgene from [65] | NK1770 |
| qIs154(lag-2p:: myr::tdTomato); lag-2(cp193[lag-2::mNeonGreen^3xFlag])V | WB Cat# JK4472, RRID:WB-STRAIN:JK4472 parent from CGC, [13] | NK2517 |
| qyIs287[myo-3p::mCherry]; lag-2(cp193[lag-2::mNeonGreen^3xFlag]) V | This study | NK2376 |
| cpIs121(lag-2p::mNG:: PLCδPH::F2A::rde-1) I; rrf-3(pk1426) II; rde-1(ne219) V | [14] | LP718 |
| cpIs121(lag-2p::mNG:: PLCδPH::F2A::rde-1) I; rrf-3(pk1426) II; sax-7(eq1) IV; rde-1(ne219) V | WB Cat# LH81, RRID:WB-STRAIN:LH81 parent from CGC, [66] | NK2035 |
| hmr-1(cp21[hmr-1::GFP+LoxP]) I; cpIs91(lag-2p::2x mKate2::PH::3xHA::tbb-2 3′UTR LoxN) II | WB Cat# LP172, RRID:WB-STRAIN:LP172 parent from [41] | NK2518 |
| cpIs91(lag-2p::2x mKate2::PH::3xHA::tbb-2 3′UTR LoxN) II; hmp-1(cp20[hmp-1::GFP + LoxP unc-119(+) LoxP]) V | WB Cat# LP191, RRID:WB-STRAIN:LP19 parent from [41] | NK2364 |
| hmp-2(cp78[GFP::hmp-2A]) I; cpIs91(lag-2p::2x mKate2::PH::3xHA::tbb-2 3′UTR LoxN) II | WB Cat# LP316, RRID:WB-STRAIN:LP316 parent from [41] | NK2368 |
| ddIs290[sax-7::TY1::EGFP::3xFLAG(92C12) + unc-119(+)] II; cpIs91[lag-2p>2x mKate2::PH::3xHA::tbb-2 3′UTR LoxN] II (heterozygotes) | WB Cat# TH502, RRID:WB-STRAIN:TH502 parent from CGC | NK2451 |
| qyIs450(myo-3p::mCherry:: PLCδPH); hmr-1(cp21[hmr-1::GFP+LoxP]) I | WB Cat# LP172, RRID:WB-STRAIN:LP172 parent from [41] | NK2173 |
| qyIs455(myo-3p::mCherry:: PLCδPH); hmp-1(cp20[hmp-1::GFP + LoxP unc-119(+) LoxP]) V | WB Cat# LP191, RRID:WB-STRAIN:LP19 parent from [41] | NK2371 |
| qyIs455(myo-3p::mCherry:: PLCδPH); hmp-2(cp78[GFP::hmp-2A]) I | WB Cat# LP316, RRID:WB-STRAIN:LP316 parent from [41] | NK2373 |
| qyIs450(myo-3p::mCherry:: PLCδPH); ddIs290[sax-7::TY1::EGFP::3xFLAG(92C12) + unc-119(+)] II | WB Cat# TH502, RRID:WB-STRAIN:TH502 parent from CGC | NK2439 |
| Oligonucleotides | ||
| myo-3 promoter F 5’-TTCCCGACAAAACATGAG-3′ | This study | N/A |
| myo-3 promoter R 5’-AGATGGATCTAGTGGTCG-3′ | This study | N/A |
|
lag-2(cp193) 5’ homology arm F 5’-CTTGCAGTGTCAAAGACGCC-3′ |
This study | N/A |
|
lag-2(cp193) 5’ homology arm R 5’-CTGGAATAGACGGTGGACTTAA-3′ |
This study | N/A |
|
lag-2(cp193) 3′ homology arm F 5’-TAGATTCTTCCTTTATCCAAAAAATGTGT-3′ |
This study | N/A |
|
lag-2(cp193) 3′ homology arm R 5’-CAATTTGGCGGCGGGATAGA-3′ |
This study | N/A |
|
lag-2(cp193) guide RNA primer 5’-GAAGAATCTAGACATAGTGAC-3′ |
This study | N/A |
| hmr-1 RNAi F 5’-ATGCAGAAGCGGCGGTGCACGT-3′ | This study | N/A |
| hmr-1 RNAi R 5’-TGAACGGAGAACACATCAAC-3′ | This study | N/A |
| Recombinant DNA | ||
| epi-1 feeding RNAi clone | Ahringer RNAi library [57] | HGMP_Location IV-5J23 |
| dgn-1 feeding RNAi clone | Ahringer RNAi library [57] | HGMP_Location X-5C20 |
| hmr-1 feeding RNAi clone | Ahringer RNAi library [57] | HGMP_Location I-5F19 |
| Improved hmr-1 feeding RNAi clone | This study | pLML30 |
| Membrane expression vector with mCherry:: PLCδPH | [67] | pAA173, Addgene plasmid #21743 |
| Membrane expression vector with GFP::CAAX | Scott Alper | pSA129 |
| Plasmid for SEC CRISPR strategy | [33] | pDD268 |
| eft-3p::Cas9+sgRNA expression vector | [33] | pDD162, Addgene plasmid #47549 |
| RNAi empty vector for new RNAi cloning and controls | Andrew Fire | L4440, Addgene plasmid #1654 |
| myo-3p::mCherry | [56] | pCFJ104 |
| unc-119 rescue plasmid | Morris Maduro | pPD#MM016B |
| Software and Algorithms | ||
| μManager software v1.4.18 | [60] | https://micro-manager.org/ |
| Axiovision software | Carl Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/axiovision.html |
| FIJI 2.0 | [68] | https://fiji.sc/ |
| Photoshop CC | Adobe Systems Inc. | N/A |
| Adobe Illustrator CC | Adobe Systems Inc. | N/A |
| FIJI stitching plug-in | [61] | https://imagej.net/Image_Stitching |
| Pic Stitch – Collage Editor | Big Blue Clip, LLC | https://itunes.apple.com/us/app/pic-stitch-collage-editor/id454768104?mt=8 |
| JMP Pro 13 | JMP®, Version 13. SAS Institute Inc., Cary, NC, 1989–2007. |
https://www.jmp.com/en_us/software/predictive-analytics-software.html |
| Imaris | Bitplane, Inc. | Versions 7 and 9 |
METHOD DETAILS
Molecular biology
We used the body wall muscle-specific expression of the myo-3 promoter to drive cytoplasmic mCherry and membrane-targeted GFP::CAAX and mCherry::PLCδPH in body wall muscle cells. A myo-3p::mCherry strain was generated by coinjecting pCFJ104 (myo-3p::mCherry) [56] with transformation marker pPD#MM016B (unc-119) as well as EcoRI-digested salmon sperm DNA and pBluescript II as carrier DNA at 50 ng/μL into unc-119(ed4) animals. A 2.3 kb region of the myo-3 promoter between forward primer 5’-TTCCCGACAAAACATGAG-3’ and reverse primer 5’-AGATGGATCTAGTGGTCG-3’ was amplified from genomic DNA by PCR and fused to GFP::CAAX (pSA129) or mCherry:: PLCδPH (pAA173) by fusion PCR and similarly injected to generate transgenic lines. pSA129 was a gift of Scott Alper. Recombinant DNA Identifiers in Key Resources Table are given as previously published or as listed on Addgene, except pLML30, which was generated for this study.
Integrated lines were obtained by standard gamma irradiation protocol.
The allele cp193(lag-2::mNeonGreen^3xFlag) was constructed using Cas9-triggered homologous recombination with a self-excising selection cassette [33]. Homology arms were amplified by PCR (5’ homology arm forward primer 5’-CTTGCAGTGTCAAAGACGCC-3’ and reverse primer 5’-CTGGAATAGACGGTGGACTTAA-3’; 3’ homology arm forward primer 5’-TAGATTCTTCCTTTATCCAAAAAATGTGT-3’ and reverse primer 5’-CAATTTGGCGGCGGGATAGA-3’) and cloned into a SpeI/AvrII-digested pDD268 backbone using Gibson assembly (New England Biolabs). Synonymous substitutions were engineered in the eft-3p::Cas9+sgRNA expression vector pDD162 using Q5 site-directed mutagenesis (New England Biolabs) and co-injected into adult germlines with repair templates and extrachromosomal array markers as described previously [32,33]. Candidate knock-ins were selected by hygromycin B treatment, phenotypic identification (roller), and the absence of fluorescent extrachromosomal arrays.
For hmr-1/cadherin RNAi, the W02B9.1 clone from the Ahringer feeding RNAi library [57] was used in the screen (Table S1) and in the experiment reported in Figure 4A. To increase efficiency of hmr-1 knockdown, a new RNAi clone targeting 1.5 kb of the N-terminus of hmr-1a was designed. An hmr-1a cDNA fragment of 1469 bp was amplified by PCR using forward primer 5’-ATGCAGAAGCGGCGGTGCACGT-3’ and reverse primer 5’-TGAACGGAGAACACATCAAC-3’ from N2 cDNA, cloned into the vector L4440, transformed into HT115 bacteria, and used in the experiment reported in Figure 4C. Effective hmr-1/cadherin knockdown by RNAi was ensured by selecting animals with body morphology defects [38] for scoring in Figure 4A, and in Figure 4C animals with visible knockdown of HMR-1::GFP fluorescence were scored.
Scoring of gonad rupture, cell enwrapment, apoptosis, and DTC processes
Gonadal ruptures were detected as breaks in the gonadal basement membrane, visible by DIC as an interrupted phase-dense line, or as animals with no visible gonadal basement membrane that were filled with germ cells. Germ cells are clearly identifiable by DIC due to their large nucleolus resulting in a distinctive “fried-egg” appearance.
To be conservative in scoring enwrapment and ensure that reduced enwrapment in the hmr-1/E-cadherin RNAi-treated animals was not caused by lack of contact between muscle and germ cells, “enwrapment” was defined as every germ cell along the dorsal body wall muscle being enwrapped, and “reduced enwrapment” was scored as half or fewer of the cells in this position being enwrapped. Muscle enwrapment of somatic gonad and coelomocytes was scored for each fluorescently highlighted cell and found to be absent in all cases (Figure 2B). Germ cell membranes were found not to be protrusive in n = 9/9 time-lapses of at least 60 minutes, defined as the germ cell membranes facing the muscle membrane not deviating in position over the time-lapse period (as in dashed line in row labeled germ cell > GFP:: PLCδPH in Figure S2.)
Apoptotic cells were identified visually by TTR-52::mCherry expression. Synchronized L1 animals were plated on OP50 and grown at 20°C for 24 hours, then transferred to epi-1 RNAi plates to induce gonad rupture and grown at 20°C for another 14 hours prior to heat-shock induction of the hsp::ttr-2::mCherry transgene at 33°C for 45 minutes (as previously performed in [25]). After recovery at 20°C for five hours, animals were imaged at the mid-L4 stage.
As a measure of germ cell enwrapment by the DTC niche, we quantified the DTC plexus by counting the number of short DTC processes intercalating between and surrounding germ cells in Fig. 4A and B, as in [13].
Germ cell counts following DTC ablation
L1 larvae were grown on standard E. coli OP50 food until the L2 stage and then transferred to epi-1 RNAi to induce gonadal ruptures. Laser ablation of the DTCs in these L3/L4 larvae was performed on a MicroPoint (Andor Technology) laser ablation setup by targeting the DTC and firing the laser until nuclear changes were detected [58]; worms were rescued back to epi-1 RNAi. The criterion for successful DTC ablation was loss of the DTC marker lag-2p::GFP::CAAX. Ablated animals and non-ablated control animals were recovered from the same slides to OP50 and imaged as adults at 48 hours post-ablation. Of these microsurgeries 4/18 ablation attempts were successful and scorable at 48 hours post ablation. To standardize germ cell counts between animals and account for variation in escaped germ cell number along the anterior-posterior axis, germ cells were counted in a 120 × 50 μm box with 15 μm z-depth centered on the ruptured gonad. Sperm were excluded from germ cell counts as sperm were typically retained within the spermatheca, which did not rupture on epi-1 RNAi.
RNAi screen
We performed a DTC-specific RNAi screen of clones targeting 2289 genes on Chromosome 1 from the Ahringer Feeding RNAi Library [57]. A strain carrying cpIs121(lag-2p:: mNG::PLCδPH::F2A::rde-1) with DTC-specific RNAi activity and fluorescently-labeled DTC was used in this screen [14]. Animals were developmentally synchronized by bleaching [59] and plated as L1 larvae and allowed to develop for ~48 hours on bacteria expressing double stranded RNA. Scoring was performed after soaking in 100 mg/ml BDM anesthetic in M9 buffer, under YFP filter fluorescence illumination on an AxioZoom V16 microscope with a PlanNeoFluar Z 2.3x/0.57 FWD 10.6 mm objective at 168x magnification. Worms were scored qualitatively for the length of the DTC as either wild-type (>50 μm), mild defect (~20–50 μm), moderate defect (~10–20 μm) or severe defect (<10 μm). For ease of analysis, wild-type and mild defects were considered unaffected, and moderate and severe defects were considered to manifest a DTC reduction phenotype (as in [14]). For the 35 clones that gave a phenotype in 9/10 or 10/10 animals examined, we verified the sequence identity of the primers given in the library, corrected any errors, and reported the correct targets in Table S1. We noted some batch effects (sequential or nearby clones tested at the same time yielding strong phenotypes). To follow up on hmr-1, we generated an improved RNAi clone (see Molecular Biology, above, and Sequential RNAi, below).
Sequential RNAi
Double RNAi against hmr-1/cadherin and epi-1/laminin was performed sequentially. A strain carrying cp21(hmr-1::GFP + LoxP) and myo-3p::mCherry::PLCδPH was used. Larval animals were transferred to hmr-1 RNAi (see Molecular Biology section for RNAi construct details), allowed to develop to reproductive age, and laid eggs on hmr-1 RNAi media. The offspring larva were allowed to develop on hmr-1 RNAi media until the late L1/early L2 stage, when they were transferred to epi-1 RNAi media to induce gonad rupture and release germ cells. Only animals with ruptured gonads were scored (see above and Figure 4C DIC images). Knockdown of hmr-1 was verified by loss or dramatic reduction of HMR-1::GFP expression in muscles and germ cells (strong expression in the uterine cells appeared to be relatively resistant to hmr-1 RNAi, as in Figure 4D).
Microscopy and image acquisition, processing, and analysis
Confocal DIC and fluorescent images were acquired on an AxioImager A1 microscope (Carl Zeiss) equipped with an EMCCD camera (Hamamatsu Photonics), a 100x or 40x Plan-Apochromat (1.4 NA) objective, and a spinning disc confocal scan head (CSU-10; Yokogawa Electric Corporation) driven by μManager software [60] at 20°C. Widefield DIC and fluorescent images were acquired with an AxioImager A1 microscope (Carl Zeiss) equipped with a CCD camera (AxioCam MRm; Carl Zeiss) and 100x Plan-Apochromat objective (1.4 NA) driven by Axiovision software (Carl Zeiss). Worms were mounted on 5% noble agar pads containing 0.01 M sodium azide for imaging during endpoint experiments. For time-lapse acquisitions, worms were anesthetized in 0.2% tricaine and 0.02% levamisole in M9. Images were processed with FIJI 2.0 and Photoshop CC (Adobe Systems Inc.). Images are displayed as single confocal z-slices or maximum intensity projections generated in FIJI. Full projections encompass the entire tissue of interest, and core projections were defined as the central confocal z-slices of the tissue. Tiled images of DTC-ablated animals in Fig. 3B were stitched using the FIJI stitching plugin [61]. Supplemental videos were generated with Imaris 7 and 9 (Bitplane) and annotated in Pic Stitch – Collage Editor. Graphs generated by JMP Pro 13 or MS Excel were refined using Illustrator CC (Adobe Systems Inc.).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification
See Experimental Methods for scoring details.
Statistical Analysis
Fisher’s exact tests, ANOVA followed by the Tukey-Kramer HSD test, or one tailed Wilcoxon Signed Rank tests were performed in JMP Pro 13 as noted in figure legends and text.
Supplementary Material
Video S1. Germ cell membranes are not invasive. Related to Figure 1 and Figure S2. Full projections through confocal z-stack of merged (top) and individual fluorescently labeled muscle cell membranes (middle, myo-3p::mCherry::PLCδPH) and germ cell membranes (bottom, mex-5p::GFP::PLCδPH). One germ cell that becomes enwrapped is marked by an asterisk. A growing muscle protrusion is marked by an arrowhead. Germ cells do not form protrusions into the muscle in n = 9/9 time-lapses of at least 60 minutes. Images were acquired every minute during a 60-minute time-lapse (same animal shown in Figure S2). Scale bar is 20 μm. Video plays at 10 fps.
Video S2: Muscle cells actively enwrap escaped germ cells. Related to Figure 1. Muscle cells (green, egl-13p::GFP) extend dynamic protrusions contacting escaped germ cells (magenta, mex-5p::H2B::mCherry) in n = 6/6 time-lapses of at least 60 minutes. Gonadal rupture was induced with epi-1/laminin RNAi treatment. A full projection of 20-μm confocal z-stacks during a 78-minute time-lapse is shown, with frames acquired every two minutes. Scale bar is 10 μm.
Video S3: Muscle cell protrusions do not form without gonadal basement membrane rupture. Related to Figure 1. In control animals treated with empty vector L4440 that have intact gonadal basement membranes and no germ cell escape, muscle cells (green, egl-13p::GFP) do not extend dynamic projections adjacent to germ cells (magenta, mex-5p::H2B::mCherry) in n = 6/6 time-lapses of at least 60 minutes. A full projection of 20-μm confocal z-stacks during a 64-minute time-lapse is shown, with frames acquired every two minutes. Scale bar is 10 μm.
Highlights.
C. elegans germ cells can escape into the body cavity after gonadal rupture.
Germ cells become selectively enwrapped by abnormally protrusive muscle cells.
This enwrapment mimics germ cell enwrapment by the intact stem cell niche.
A common suite of adhesion proteins mediates enwrapment in both contexts.
Acknowledgements
We thank Geraldine Seydoux and members of the Sherwood lab for discussions and Jeremy Nance for providing strains. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Agnieszka Kawska (info@illuscientia) for her work on schematics and Laura Kelley (Duke University) for her help with video editing. We thank Jonnathan Singh Alvarado (Duke University) for experimental assistance. K.L.G. was supported by postdoctoral fellowship GM121015–01 from the National Institutes of Health. L.M.L. was supported by graduate research fellowship DGE-1106401 from the National Science Foundation. A.M.P. was supported by postdoctoral fellowship F32 GM115151 from the National Institutes of Health and postdoctoral fellowship PF-16–030 DDC from the American Cancer Society. This work was supported by NIGMS R01 GM083071 to B.G., NIGMS R01 GM61706 to E.J.A.H., and NIGMS R01 GM079320 and NIGMS R35 MIRA GM118049 to D.R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Overholtzer M, and Brugge JS (2008). The cell biology of cell-in-cell structures. Nat. Rev. Mol. Cell Biol 9, 796–809. [DOI] [PubMed] [Google Scholar]
- 2.Nourshargh S, Hordijk PL, and Sixt M (2010). Breaching multiple barriers: leukocyte motility through venular walls and the interstitium. Nat. Rev. Mol. Cell Biol 11, 366–378. [DOI] [PubMed] [Google Scholar]
- 3.Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, Podbilewicz B, Wang H, and Wong M (2013). Genetic basis of cell–cell fusion mechanisms. Trends Genet. 29, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley LC, Lohmer LL, Hagedorn EJ, and Sherwood DR (2014). Traversing the basement membrane in vivo: a diversity of strategies. J. Cell Biol 204, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noubissi FK, and Ogle BM (2016). Cancer cell fusion: Mechanisms slowly unravel. Int. J. Mol. Sci 17, 1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu T-L, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, et al. (2018). Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360, eaaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oatley JM, and Brinster RL (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev 92, 577–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathur D, Bost A, Driver I, and Ohlstein B (2010). A transient niche regulates the specification of Drosophila intestinal stem cells. Science 327, 210–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins AB, McCaffery JM, and Van Doren M (2003). Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development 130, 4417–26. [DOI] [PubMed] [Google Scholar]
- 10.Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, Yzaguirre AD, Speck NA, and Zon LI (2015). Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohrschneider MR, and Nance J (2013). The union of somatic gonad precursors and primordial germ cells during Caenorhabditis elegans embryogenesis. Dev. Biol 379, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, and Greenstein D (1999). Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: Relations between the germ line and soma. Dev. Biol 212, 101–123. [DOI] [PubMed] [Google Scholar]
- 13.Byrd DT, Knobel K, Affeldt K, Crittenden SL, and Kimble J (2014). A DTC niche plexus surrounds the germline stem cell pool in Caenorhabditis elegans. PLoS One 9, e88372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linden LM, Gordon KL, Pani AM, Payne SG, Garde A, Burkholder D, Chi Q, Goldstein B, and Sherwood DR (2017). Identification of regulators of germ stem cell enwrapment by its niche in C. elegans. Dev. Biol 429, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inaba M, Yamashita YM, and Buszczak M (2016). Keeping stem cells under control: New insights into the mechanisms that limit niche-stem cell signaling within the reproductive system. Mol. Reprod. Dev 83, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baum PD, and Garriga G (1997). Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron 19, 51–62. [DOI] [PubMed] [Google Scholar]
- 17.Huang C-C, Hall DH, Hedgecock EM, Kao G, Karantza V, Vogel BE, Hutter H, Chisholm AD, Yurchenco PD, and Wadsworth WG (2003). Laminin alpha subunits and their role in C. elegans development. Development 130, 3343–58. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RP, Kang SH, and Kramer JM (2006). C. elegans dystroglycan DGN-1 functions in epithelia and neurons, but not muscle, and independently of dystrophin. Development 133, 1911–21. [DOI] [PubMed] [Google Scholar]
- 19.Trzebiatowska A, Topf U, Sauder U, Drabikowski K, and Chiquet-Ehrismann R (2008). Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol. Biol. Cell 19, 3898–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi W, Huang X, Neumann-Haefelin E, Schulze E, and Baumeister R (2012). Cell-nonautonomous signaling of FOXO/DAF-16 to the stem cells of Caenorhabditis elegans. PLoS Genet. 8, e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. (2018). Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidel HS, Smith TA, Evans JK, Stamper JQ, Mast TG, and Kimble J (2018). C. elegans germ cells divide and differentiate in a folded tissue. Dev. Biol 442, 173–187. [DOI] [PubMed] [Google Scholar]
- 23.Kadyk LC, and Kimble J (1998). Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125, 1803–1813. [DOI] [PubMed] [Google Scholar]
- 24.Altun ZF, and Hall DH (2009). Coelomocyte system. Wormatlas. Available at: http://www.wormatlas.org/hermaphrodite/coelomocyte/Coelomoframeset.html.
- 25.Wang X, Li W, Zhao D, Liu B, Shi Y, Chen B, Yang H, Guo P, Geng X, Shang Z, et al. (2010). Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell Biol 12, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander M, Chan KKM, Byrne AB, Selman G, Lee T, Ono J, Wong E, Puckrin R, Dixon SJ, and Roy PJ (2009). An UNC-40 pathway directs postsynaptic membrane extension in Caenorhabditis elegans. Development 136. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, Hartwieg E, and Horvitz HR (2001). CED-1 Is a Transmembrane Receptor that Mediates Cell Corpse Engulfment in C. elegans. Cell 104, 43–56. [DOI] [PubMed] [Google Scholar]
- 28.Berry LW, Westlund B, and Schedl T (1997). Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124, 925–936. [DOI] [PubMed] [Google Scholar]
- 29.Henderson ST, Gao D, Lambie EJ, and Kimble J (1994). lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913–24. [DOI] [PubMed] [Google Scholar]
- 30.Fitzgerald K, and Greenwald I (1995). Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development 121, 4275–82. [DOI] [PubMed] [Google Scholar]
- 31.Gao D, and Kimble J (1995). APX-1 can substitute for its homolog LAG-2 to direct cell interactions throughout Caenorhabditis elegans development. Proc. Natl. Acad. Sci. U. S. A 92, 9839–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, and Goldstein B (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambie EJ, and Kimble J (1991). Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112. [DOI] [PubMed] [Google Scholar]
- 35.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, and Banerjee U (2007). A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature 446, 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith NR, Davies PS, Silk AD, and Wong MH (2012). Epithelial and mesenchymal contribution to the niche: A safeguard for intestinal stem cell homeostasis. Gastroenterology 143, 1426–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blazie SM, Babb C, Wilky H, Rawls A, Park JG, and Mangone M (2015). Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol. 13, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa M, Raich W, Agbunag C, Leung B, Hardin J, and Priess JR (1998). A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol 141, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Doren M, Mathews WR, Samuels M, Moore LA, Broihier HT, and Lehmann R (2003). fear of intimacy encodes a novel transmembrane protein required for gonad morphogenesis in Drosophila. Development 130, 2355–64. [DOI] [PubMed] [Google Scholar]
- 40.Grana TM, Cox EA, Lynch AM, and Hardin J (2010). SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev. Biol 344, 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marston DJ, Higgins CD, Peters KA, Cupp TD, Dickinson DJ, Pani AM, Moore RP, Cox AH, Kiehart DP, and Goldstein B (2016). MRCK-1 drives apical constriction in C. elegans by linking developmental patterning to force generation. Curr. Biol 26, 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei CH, and Ryu SE (2012). Homophilic interaction of the L1 family of cell adhesion molecules. Exp. Mol. Med 44, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim B, and Emmons SW (2017). Multiple conserved cell adhesion protein interactions mediate neural wiring of a sensory circuit in C. elegans. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L, Ong B, and Bennett V (2001). LAD-1, the Caenorhabditis elegans L1CAM homologue, participates in embryonic and gonadal morphogenesis and is a substrate for fibroblast growth factor receptor pathway-dependent phosphotyrosine-based signaling. J. Cell Biol 154, 841–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merritt C, and Seydoux G (2010). Transgenic solutions for the germline. WormBook, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, and Schulz C (2007). Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr. Biol 17, 1253–1258. [DOI] [PubMed] [Google Scholar]
- 47.Kimble JE, and White JG (1981). On the control of germ cell development in Caenorhabditis elegans. Dev. Biol 81, 208–219. [DOI] [PubMed] [Google Scholar]
- 48.Kai T, and Spradling A (2003). An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc. Natl. Acad. Sci 100, 4633–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitadate Y, and Kobayashi S (2010). Notch and Egfr signaling act antagonistically to regulate germ-line stem cell niche formation in Drosophila male embryonic gonads. Proc. Natl. Acad. Sci. U. S. A 107, 14241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hétié P, de Cuevas M, and Matunis E (2014). Conversion of quiescent niche cells to somatic stem cells causes ectopic niche formation in the Drosophila testis. Cell Rep. 7, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu R, Li J, Zhao H, Kong R, Wei M, Shi L, Bai G, and Li Z (2018). Self-restrained regulation of stem cell niche activity by niche components in the Drosophila testis. Dev. Biol 439, 42–51. [DOI] [PubMed] [Google Scholar]
- 52.McGovern M, Voutev R, Maciejowski J, Corsi AK, and Hubbard EJA (2009). A “latent niche” mechanism for tumor initiation. Proc. Natl. Acad. Sci. U. S. A 106, 11617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sirka OK, Shamir ER, and Ewald AJ (2018). Myoepithelial cells are a dynamic barrier to epithelial dissemination. J. Cell Biol 217, 3368–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Psaila B, and Lyden D (2009). The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, et al. (2017). Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317. [DOI] [PubMed] [Google Scholar]
- 56.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, and Jorgensen EM (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet 40, 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamath RS, and Ahringer J (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321. [DOI] [PubMed] [Google Scholar]
- 58.Bargmann CI, and Avery L (1995). Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48, 225–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stiernagle T (2006). Maintenance of C. elegans. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edelstein A, Amodaj N, Hoover K, Vale R, and Stuurman N (2010). Computer control of microscopes using manager. Curr. Protoc. Mol. Biol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Preibisch S, Saalfeld S, and Tomancak P (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bourdages KG, Lacroix B, Dorn JF, Descovich CP, and Maddox AS (2014). Quantitative analysis of cytokinesis in situ during C. elegans postembryonic development. PLoS One 9, e110689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chihara D, and Nance J (2012). An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation. Development 139, 2547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayes GD, Frand AR, and Ruvkun G (2006). The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133, 4631–4641. [DOI] [PubMed] [Google Scholar]
- 65.Roy D, Kahler DJ, Yun C, and Hubbard EJA (2018). Functional interactions between rsks-1/S6K, glp-1/Notch, and regulators of Caenorhabditis elegans fertility and germline stem cell maintenance. G3 (Bethesda). 8, 3293–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou S, Opperman K, Wang X, and Chen L (2008). unc-44 Ankyrin and stn-2 y-Syntrophin Regulate sax-7 L1CAM Function in Maintaining Neuronal Positioning in Caenorhabditis elegans. Genetics 180, 1429–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziel JW, Hagedorn EJ, Audhya A, and Sherwood DR (2009). UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat. Cell Biol 11, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Germ cell membranes are not invasive. Related to Figure 1 and Figure S2. Full projections through confocal z-stack of merged (top) and individual fluorescently labeled muscle cell membranes (middle, myo-3p::mCherry::PLCδPH) and germ cell membranes (bottom, mex-5p::GFP::PLCδPH). One germ cell that becomes enwrapped is marked by an asterisk. A growing muscle protrusion is marked by an arrowhead. Germ cells do not form protrusions into the muscle in n = 9/9 time-lapses of at least 60 minutes. Images were acquired every minute during a 60-minute time-lapse (same animal shown in Figure S2). Scale bar is 20 μm. Video plays at 10 fps.
Video S2: Muscle cells actively enwrap escaped germ cells. Related to Figure 1. Muscle cells (green, egl-13p::GFP) extend dynamic protrusions contacting escaped germ cells (magenta, mex-5p::H2B::mCherry) in n = 6/6 time-lapses of at least 60 minutes. Gonadal rupture was induced with epi-1/laminin RNAi treatment. A full projection of 20-μm confocal z-stacks during a 78-minute time-lapse is shown, with frames acquired every two minutes. Scale bar is 10 μm.
Video S3: Muscle cell protrusions do not form without gonadal basement membrane rupture. Related to Figure 1. In control animals treated with empty vector L4440 that have intact gonadal basement membranes and no germ cell escape, muscle cells (green, egl-13p::GFP) do not extend dynamic projections adjacent to germ cells (magenta, mex-5p::H2B::mCherry) in n = 6/6 time-lapses of at least 60 minutes. A full projection of 20-μm confocal z-stacks during a 64-minute time-lapse is shown, with frames acquired every two minutes. Scale bar is 10 μm.