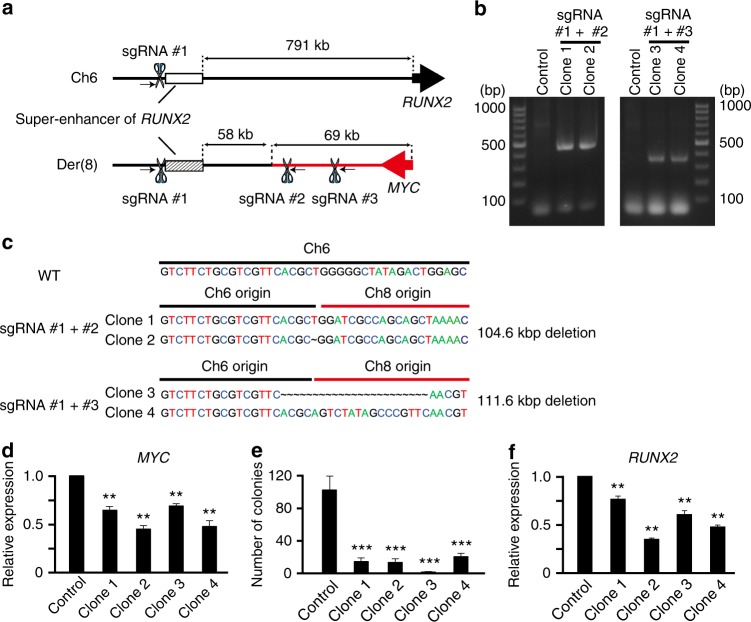
Fig. 5.
RUNX2 super-enhancer-driven MYC expression promotes the formation of BPDCN. a Schematic illustration of the RUNX2 super-enhancers on chromosome 6 (seRUNX2 (-791kb); a back line) and derivative chromosome 8 (seRUNX2der8; a black and red fused line). sgRNA vectors were directed a distal region of the super-enhancer (sgRNA #1; chr6: 44,557,984-44,558,006) and two distinct downstream regions of MYC (sgRNA #2; chr8:128,812,545-128,812,567 and sgRNA #3; chr8:128,805,565-128,805,587). Arrows indicate primers for genomic PCR utilized in Fig. 5b. b Detecting successful deletions of the seRUNX2der8 by genomic PCR using primers on distinct chromosomes of 6 and 8. c Sequences of seRUNX2der8-deleted clones elucidated by Sanger sequencing and sizes of deletion in those clones. d Expression levels of MYC mRNA in seRUNX2der8-deleted CAL-1 clones examined by q-PCR. e Impaired colony formation capacities in seRUNX2der8-deleted CAL-1 clones from those in control cells (n = 3). f Expression levels of RUNX2 mRNA in seRUNX2der8-deleted CAL-1 clones examined by q-PCR. d–f Bars show the mean±SD, **p < 0.01 and ***p < 0.001 by the Student’s t-test