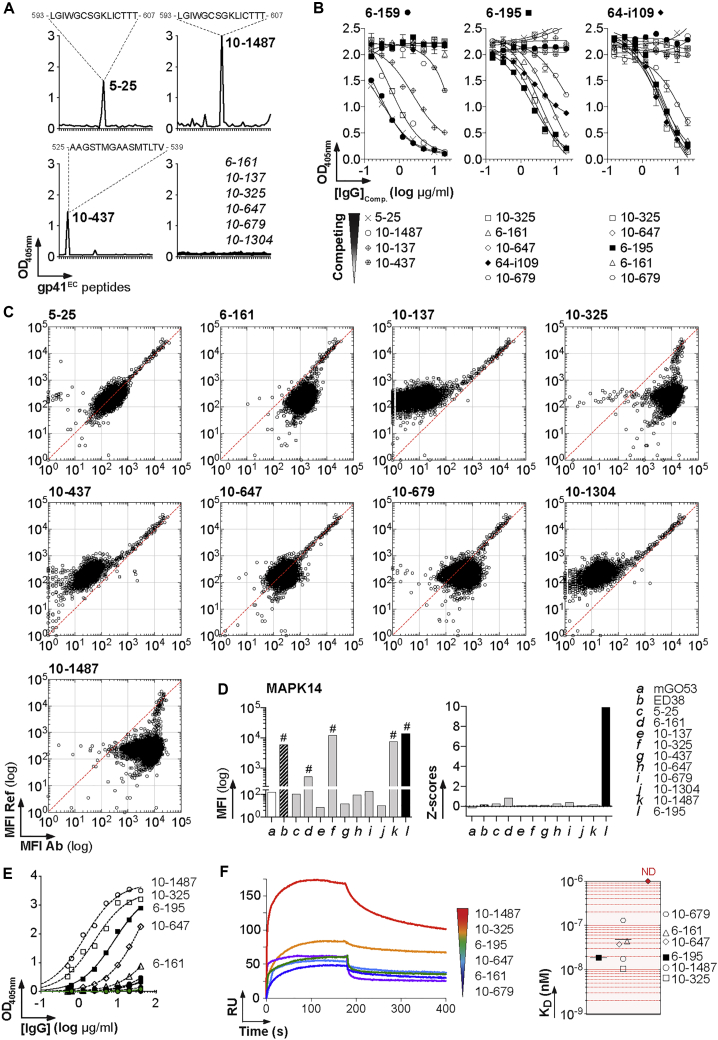
Figure 5.
Binding of HIV-1 gp41 HR2 Antibodies to MAPK14
(A) Representative ELISA graphs show the reactivity of blood anti-gp41 antibodies against overlapping peptides covering the extracellular portion of gp41 (gp41EC). The sequences of the peptides recognized are indicated on the top.
(B) Representative ELISA graphs show the binding of biotinylated mucosal anti-gp41 to YU-2 gp140 in the presence of potential competitor antibodies (blood-derived anti-gp41 IgG). Error bars indicate the SEM of duplicate values.
(C) Protein microarray plots show the reactivity profile of blood anti-gp140 antibodies against human proteins. For each protein spot, the mean fluorescence intensity (MFI) given by the reference (mGO53) and test antibody are depicted on the y and x axes, respectively. Each dot represents the average of duplicate array proteins. The diagonal lines indicate equal binding for reference and test antibodies.
(D) Bar graphs comparing the MFI values (Left) and Z scores (Right) for the binding of the selected anti-gp41 antibodies to MAPK14 immobilized on the protein array.
(E) Representative ELISA graph comparing the reactivity of blood anti-gp140 antibodies to purified MAPK14 proteins. The 6-195 and mGO53 (green) were used as positive and negative control, respectively. Error bars indicate the SEM of duplicate values.
(F) Affinity of anti-gp41 antibodies to MAPK14. (Left) Representative SPR curves comparing the binding overtime of selected anti-gp41 IgGs (at 250 nM) to purified MAPK14 immobilized on the sensor chip (250 RUs). (Right) Dot plots comparing the relative affinity (KD) of anti-gp41 antibodies to MAPK14. No binding could be detected (ND) for the following antibodies: 5-25, 10-137, 10-437, and 10-1304.
See also Figure S6.