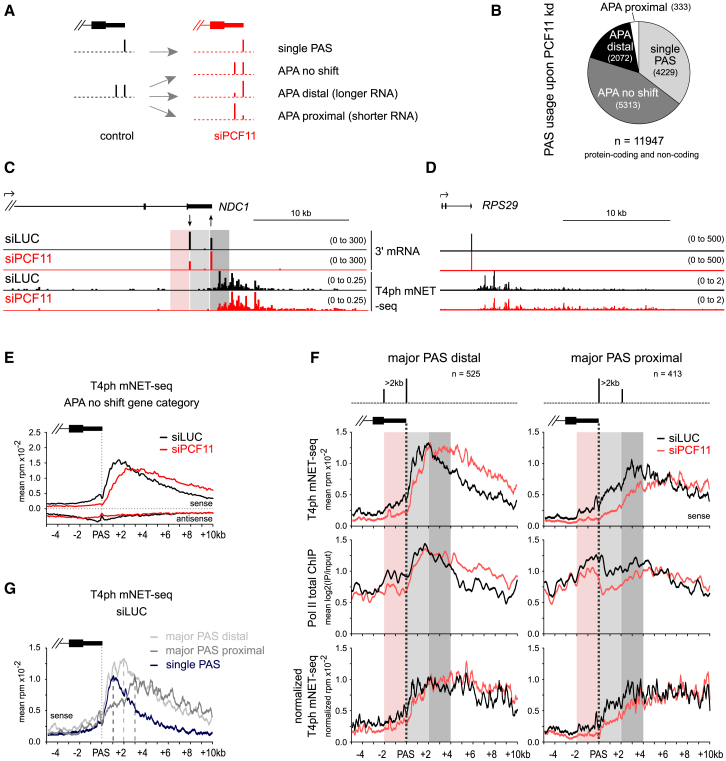
Figure 2.
PCF11-Mediated Termination Enhancement Occurs Independently of PAS Selection
(A) Schematic of gene type based on APA changes ± siPCF11.
(B) Pie chart of PAS usage in cells depleted of PCF11 based on DEXseq analysis (padj < 0.05).
(C–D) Genomic profiles of NDC1 (C) and RPS29 (D). Arrows indicate significant APA upon PCF11 depletion (DEXseq padj < 0.05; Figure S2B).
(E–G) Meta-gene profiles of T4ph mNET-seq signal around major PAS on pc genes. Vertical dotted line: position of the major PAS.
(E) Multiple PAS-containing genes without significant change in PAS usage (APA no shift); meta-profiles for other APA gene categories shown in Figure S2E.
(F) Genes with two strongest PASs of comparable signal and separated by >2 kb (n = 938) were divided into two sets: those with a major distal (left panels) or proximal (right panels) PAS. Schemes of the PAS positioning are shown on top. Top panels: T4ph mNET-seq; middle panels: Pol II total ChIP-seq; bottom panel: T4ph mNET-seq normalized to Pol II total. Red shading highlights region 2 kb upstream of major PAS, light gray 2 kb downstream, and dark gray 2–4 kb downstream of major PAS (also in C).
(G) T4ph mNET-seq profiles in control cells (siLUC) for the indicated gene categories. Vertical dashed lines highlight corresponding T4ph mNET-seq signal maxima.