Figure 4.
Closely Spaced Genes Are PCF11 Dependent
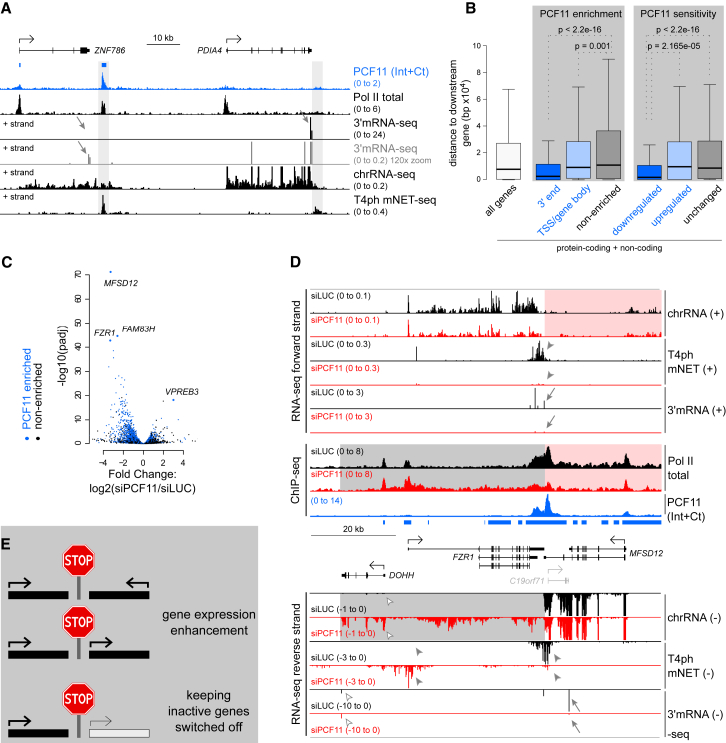
(A) Genomic profile of ZNF786/PDIA4. Blue bars indicate PCF11 enrichment. Termination regions are shaded. 3′ mRNA-seq data are shown at two viewing ranges.
(B) Boxplot of distances between gene PAS to their nearest gene downstream. Statistical significance was determined using Mann-Whitney test. In all boxplot figures, the thick horizontal line marks median and the upper and lower limits of the box the 1st and 3rd quartile.
(C) Volcano plot showing differential expression ± siPCF11. Blue dots correspond to PCF11-enriched genes, and black dots correspond to non-enriched genes. Most significantly deregulated genes are indicated.
(D) Genomic profile of the FZR1/MFSD12 locus. Data from the + strand and non-strand specific data are shown above the locus; data from the – strand are below it. Gray shading: readthrough of MFSD12, red shading: lack of detectable readthrough from FZR1. FZR1 is less active (10× zoomed in viewing range). Blue bars: PCF11 enrichment; arrows: gene downregulation; filled arrowheads: alterations in T4ph mNET-seq signal; empty arrowheads: upregulation of DOHH due to readthrough from MFSD12.
(E) Model of PCF11 role in enhancing gene expression of closely spaced genes and isolating inactive genes from upstream tandem active genes.