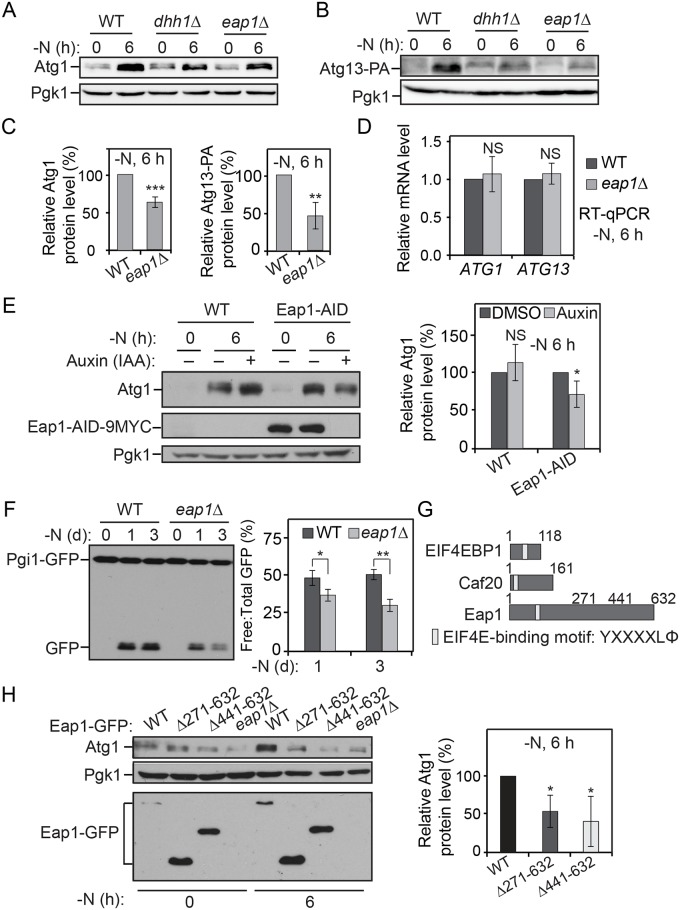
Fig 6. Eap1 facilitates Atg1 and Atg13 translation during nitrogen starvation.
(A, B, C) Atg13–PA (ZYY202) and Atg13–PA eap1Δ (ZYY204) cells were grown in YPD to mid-log phase (-N, 0 h) and then shifted to SD-N for 6 h. Cell lysates were prepared, subjected to SDS-PAGE, and analyzed by western blot. Atg1 and Atg13–PA levels are shown in (A) and (B), respectively. The quantification of Atg1 and Atg13–PA protein levels was conducted as indicated in Fig 2A and was shown in (C). **p < 0.01. ***p < 0.001. (D) Atg13–PA (ZYY202) and Atg13–PA eap1Δ (ZYY204) cells were grown in YPD to mid-log phase (-N, 0 h) and then shifted to SD-N for 6 h. Total RNA for each sample was extracted, and the ATG1 and ATG13 mRNA levels were quantified by RT-qPCR. (E) WT (XLY338) and Eap1–AID (ZYY210) cells were grown in YPD to mid-log phase and treated with either DMSO or 300 μM IAA for 30 min. They were then shifted to SD-N for 6 h in the presence of either DMSO or IAA. Cell lysates were prepared, subjected to SDS-PAGE and analyzed by western blot. The quantification of Atg1 levels was conducted as indicated in Fig 2D. *p < 0.05. (F) Pgi1–GFP (XLY306) and Pgi1–GFP eap1Δ (XLY310) cells were grown in YPD to mid-log phase (-N, 0 d) and then shifted to SD-N for 1 or 3 d. Cell lysates were prepared, subjected to SDS-PAGE, and analyzed by western blot. The quantification of the ratio of free to total GFP was conducted as indicated in Fig 1C. *p < 0.05. **p < 0.01. (G) Schematic images of the domains of EIF4EBPs, including EIF4EBP1 (Homo sapiens), Caf20 (Saccharomyces cerevisiae), and Eap1 (S. cerevisiae). Although they share an EIF4E binding motif, Eap1 has a long C-terminal stretch that is absent in the other two proteins. (H) WT Eap1–GFP (ZYY215), Eap1Δ271–632–GFP (ZYY211), Eap1Δ441–632–GFP (ZYY212), and eap1Δ (ZYY204) cells were grown in YPD to mid-log phase (-N, 0 d) and then shifted to SD-N for 6 h. Cell lysates were prepared, subjected to SDS-PAGE, and analyzed by western blot. The quantification of Atg1 protein levels was conducted as indicated in Fig 2A. *p < 0.05. (See also S7 Fig; raw numerical values are shown in S1 Data). AID, auxin-inducible degron; Atg, autophagy-related; Caf20, cap associated factor 20; DMSO, dimethyl sulfoxide; Eap1, EIF4E–associated protein 1; EIF4E, eukaryotic translation initiation factor 4E; EIF4EBP, EIF4E binding protein; GFP, green fluorescent protein; IAA, indole-3-acetic acid; NS, not significant; PA, protein A; Pgi1, phosphoglucoisomerase 1; Pgk1, 3-phosphoglycerate kinase 1; RT-qPCR, quantitative reverse transcription PCR; SD-N, synthetic minimal medium lacking nitrogen; WT, wild type; YPD, yeast extract–peptone–dextrose.