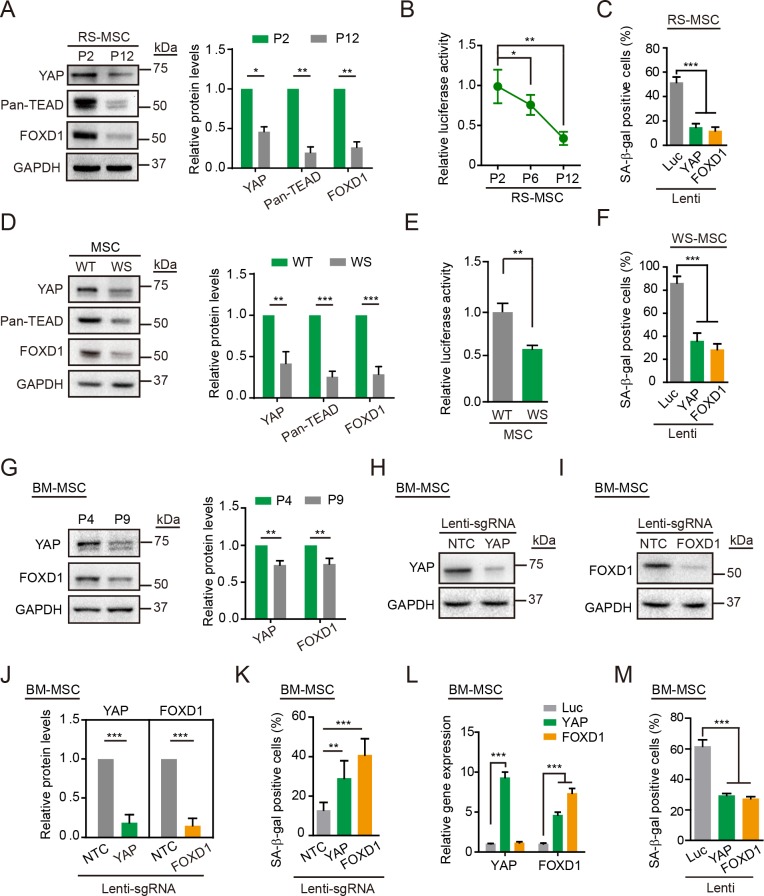
Fig 5. The YAP–FOXD1 axis counteracts replicative and pathological senescence.
(A) Western blot analysis of YAP, Pan-TEAD, and FOXD1 in RS hMSCs. GAPDH was used as a loading control. The protein levels normalized with GAPDH were shown as fold change relative to P2 hMSCs (right). Data are presented as the mean ± SD, n = 3, *P < 0.05, **P < 0.01. (B) The 8 × GTIIC-Luc activity detected in RS hMSCs. Data are presented as the mean ± SD, n = 3, *P < 0.05, **P < 0.01. (C) SA-β-gal staining of RS hMSCs transduced with lentiviruses expressing Luc, YAP, or FOXD1. Data are presented as the mean ± SD, n = 3, ***P < 0.001. (D) Western blot analysis of YAP, Pan-TEAD, and FOXD1 in WT and WS hMSCs. GAPDH was used as a loading control (left). The protein levels normalized with GAPDH were shown as fold change relative to WT hMSCs (right). Data are presented as the mean ± SD, n = 3, **P < 0.01, ***P < 0.001. (E) The 8 × GTIIC-Luc activity determined in WT and WS hMSCs. Data are presented as the mean ± SD, n = 3, **P < 0.01. (F) SA-β-gal staining of WS hMSCs transduced with lentiviruses expressing Luc, YAP, or FOXD1. Data are presented as the mean ± SD, n = 3, ***P < 0.001. (G) Western blot analysis of YAP and FOXD1 in RS BM-hMSCs. GAPDH was used as a loading control (left). The protein levels normalized with GAPDH were shown as fold change relative to P4 BM-hMSCs (right). Data are presented as the mean ± SD, n = 3, **P < 0.01. (H) Western blot analysis of YAP in BM-hMSCs transduced with lentiviruses expressing NTC or YAP sgRNA as well as CRISPR/Cas9. GAPDH was used as a loading control. (I) Western blot analysis of FOXD1 in BM-hMSCs transduced with lentiviruses expressing NTC or FOXD1 sgRNA. GAPDH was used as a loading control. (J) The protein levels normalized with GAPDH were shown as fold change relative to lenti-NTC sgRNA transduced BM-hMSCs. Data are presented as the mean ± SD, n = 3, ***P < 0.001. (K) SA-β-gal staining of BM-hMSCs transduced with lentiviruses expressing NTC, YAP, or FOXD1 sgRNA as well as CRISPR/Cas9. Data are presented as the mean ± SD, n = 3, **P < 0.01, ***P < 0.001. (L) RT-qPCR detection of YAP and FOXD1 expression levels in BM-hMSCs transduced with lentiviruses expressing Luc, YAP, or FOXD1. Data are presented as the mean ± SD, n = 3, ***P < 0.001. (M) SA-β-gal staining of BM-hMSCs transduced with lentiviruses expressing Luc, YAP, or FOXD1. Data are presented as the mean ± SD, n = 3, ***P < 0.001. The numerical data underlying this figure are included in S8 Data. BM-hMSC, human mesenchymal stem cells isolated from human bone marrow; Cas9, CRISPR associated protein 9 nuclease; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; FOXD1, forkhead box D1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; hMSC, human mesenchymal stem cell; lenti, lentivirus; Luc, luciferase; ns, not significant; NTC, non-targeting control; RS, replicative-senescent; RT-qPCR, reverse transcription quantitative polymerase chain reaction; SA-β-gal, senescence-associated-β-galactosidase; sgRNA, single guide RNA; TEAD, TEA domain transcriptional factor; WS, Werner syndrome; WT, wild type; YAP, Yes-associated protein.