Figure 4.
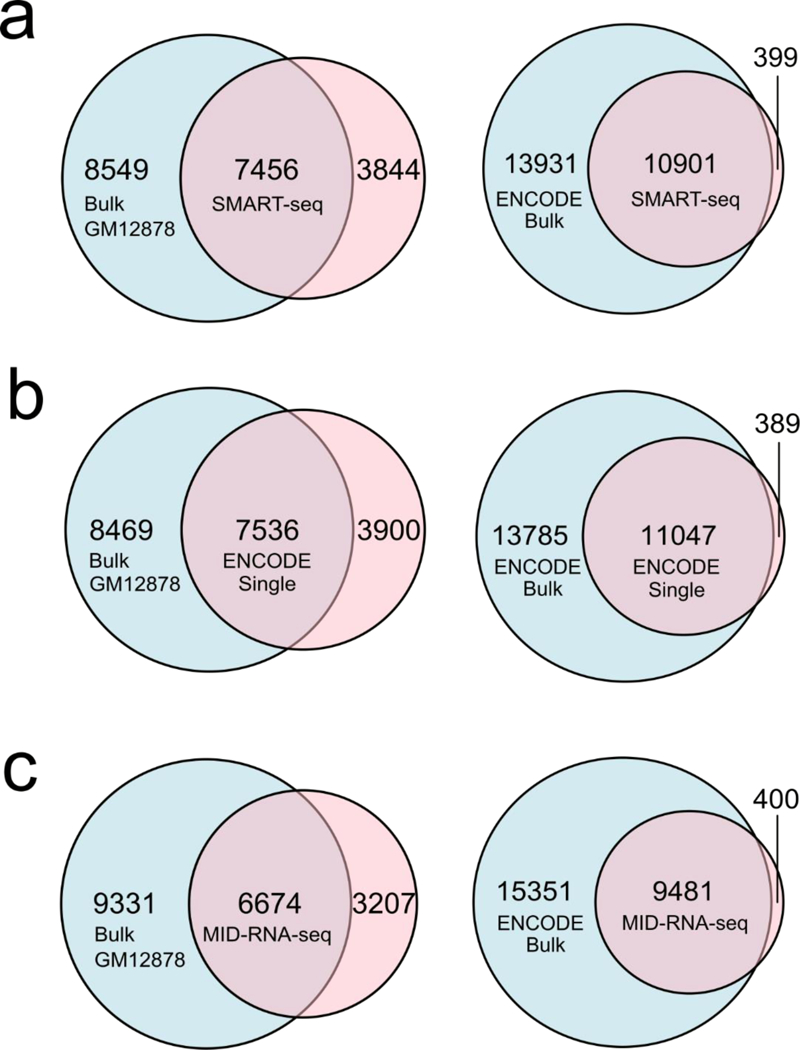
Sensitivity of various scRNA-seq techniques measured by overlap of genes between the single-cell data and the bulk RNA-seq data on GM12878 cell line. ENCODE bulk was produced using 100ng of mRNA from GM12878 cells (GSE33480) and Bulk GM12878 was produced in our lab using 1000 cells and SMART-seq2 protocol. Sequencing depth was adjusted to 2 million reads for each data set for comparison. a) SMART-seq single cell data (n=15); b) ENCODE single-cell data (n=31); c) MID-RNA-seq single-cell data (n=35). ENCODE single cell data was obtained from GSM2343071/2, SMART-seq from Ref.1.
