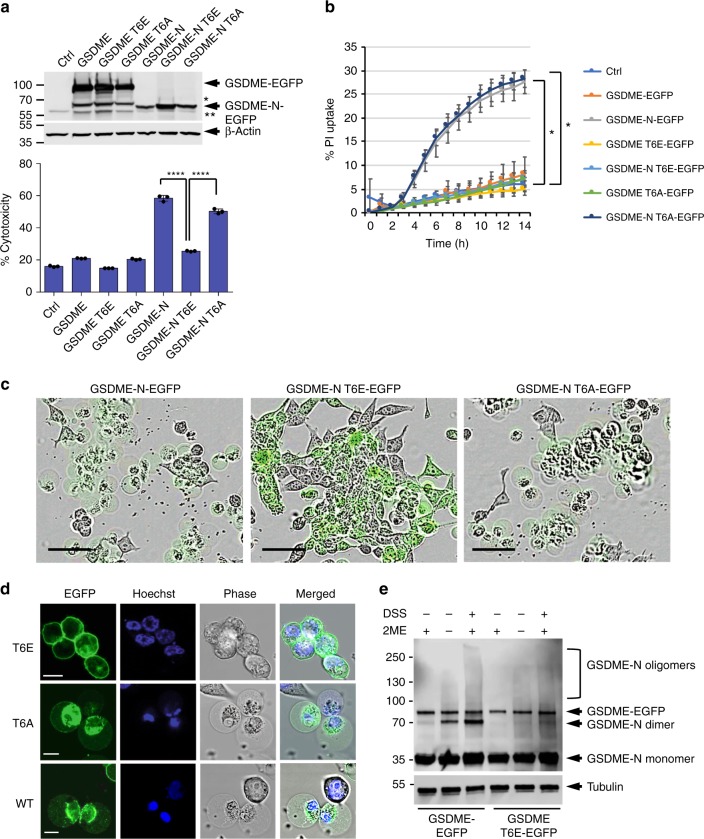
Fig. 2.
T6E mutation inhibits the pyroptotic and self-oligomerization activities of GSDME. a, b Cytotoxicity and pyroptotic activity of the indicated GSDME-N constructs as measured by lactate dehydrogenase (LDH) release (a) or propidium iodide (PI) uptake (b), respectively, in 293T cells transfected with empty vector (Ctrl) or the indicated GSDME. Expression of these constructs in cell lysates is shown in (a, top panel) as visualized by immunoblot analysis with anti-GSDME antibody. *Internal translation. **Non-specific band. c Representative IncuCyte images showing pyroptosis induction in cells expressing GSDME-N-EGFP (left) or GSDME-N-T6A-EGFP (right), but not in cells expressing GSDME-N-T6E-EGFP (middle). Scale bar, 50 µm. d Confocal images of GSDME-N-EGFP variants T6E (top panels), T6A (middle panels) and wild-type (WT; lower panels) transiently expressed in 293T cells. The perinuclear localization of WT or T6A GSDME-EGFP is largely due to the association of GSDME-N-EGFP with the mitochondria (see Fig. 4), which coalesce around the nucleus in pyroptotic cells. Scale bar, 10 µm. e Immunoblots showing WT or T6E GSDME-N after incubation with purified cell membranes and fractionation on sodium dodecyl sulfate (SDS) polyacrylamide gel in the presence or absence of β-mercaptoethanol (2-ME), or incubation with purified cell membranes followed by cross-linking with disuccinimidyl suberate (DSS) and fractionation on SDS–polyacrylamide gel in the presence of 2-ME. Results are representative of at least three independent experiments performed in duplicate or triplicate. Error bars represent S.D. Student’s t-test, *p < 0.05, ****p < 0.00005