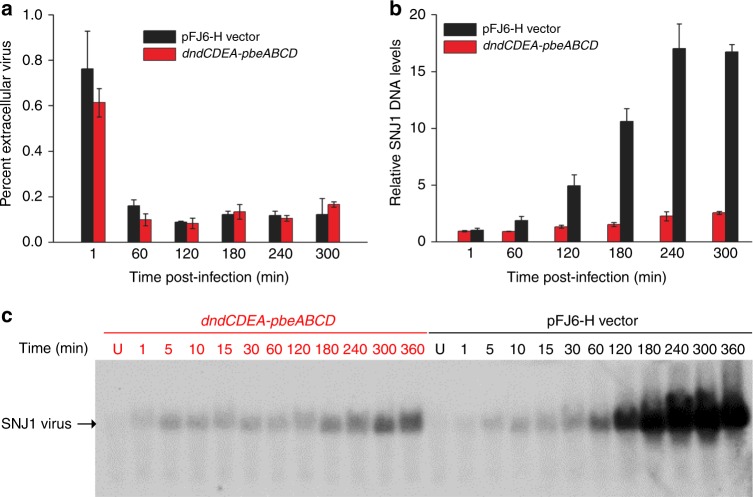
Fig. 5.
Adsorption and DNA replication of the SNJ1 virus in CJ7-F cells expressing or lacking DndCDEA-PbeABCD. a Adsorption of SNJ1 to dndCDEA-pbeABCD-containing CJ7-F cells (red) is not impaired compared to that observed for CJ7-F cells containing the empty vector pFJ6-H (black). After the infection of logarithmic-stage cultures (OD600 = 0.3) with SNJ1 at an MOI = 1, samples were collected at 60-min intervals, and the extracellular (unabsorbed) viral concentration was measured and compared to the initial viral concentrations. The bars represent the means of three experiments, and the error bars represent the SD of the mean. b Real-time qPCR analysis to determine the replication efficiency of the virus SNJ1 in CJ7-F cells expressing (red) or lacking (black) dndCDEA-pbeABCD. Primers amplifying repA and radA were used to determine the concentration of SNJ1 DNA and CJ7-F chromosomes, respectively. The 2CT(radA) – CT(repA) method was used to determine the relative levels of SNJ1 DNA in CJ7-F hosts. c Southern blot analysis of the SNJ1 genome during the infection cycle. Numbers indicate the time (min) following infection. A probe was designed corresponding to positions 9243–9732 nt in the SNJ1 genome. Source data are provided as a Source Data file