Figure 1.
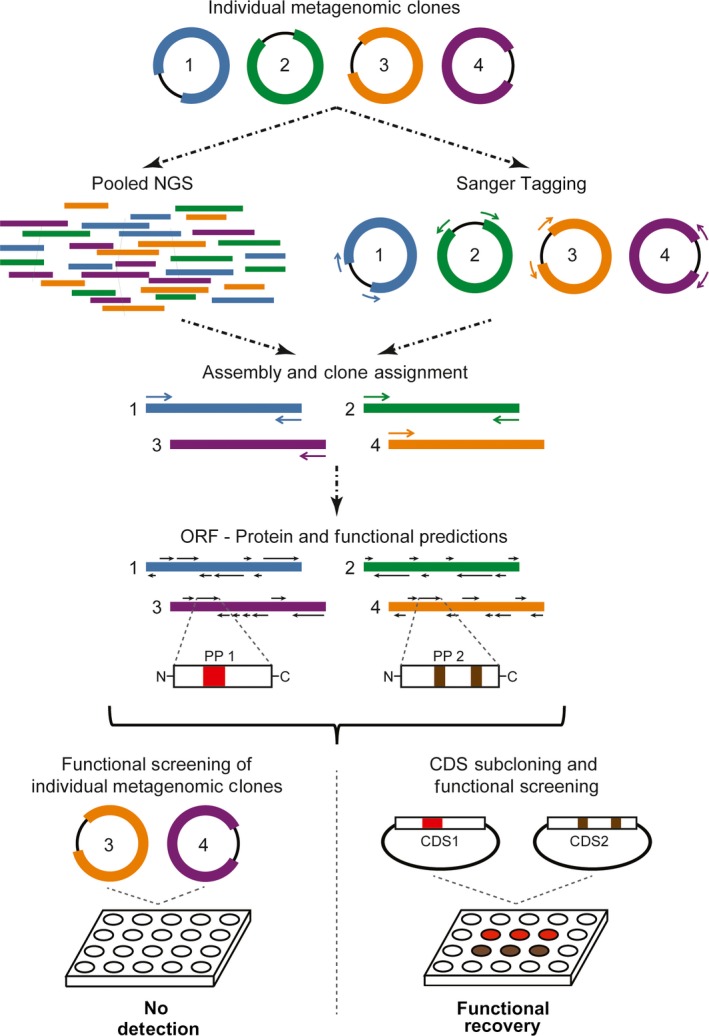
Pipeline overview. Isolated metagenomic clones are pooled in one sample for a massive sequencing analysis and independently analyzed by Sanger sequencing, in order to map the metagenomic inserts to their corresponding bacterial clones. After DNA assembly and clone assignment processes, ORF predictions and functional characterization of predicted putative proteins (e.g., PP1 and PP2) are performed. Selected coding sequences (e.g., CDS1 and CDS2) associated with the enzymatic activities of interest are matched to the original metagenomic clones or subcloned for independent maintenance in plasmid vectors. Finally, functional analyses on subclones expressing the predicted proteins allow the recovery of several enzymatic activities not identified in traditional functional metagenomic assays
