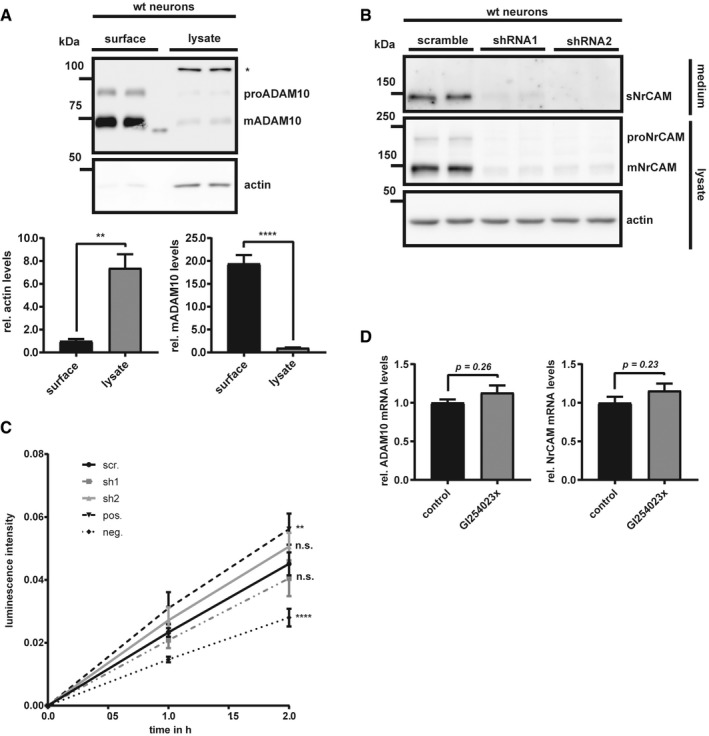
Figure EV3. Surface biotinylation efficiency and knock‐down of NrCAM using shRNA‐containing lentiviruses.
-
ATo test the efficacy of our biotinylation assay, 100 μg of total protein was used for streptavidin pull‐down, run next to 10 μg of total lysate, and were compared for the respective abundance of β‐actin and ADAM10. Densitometric quantifications of the Western blots are shown. Two‐sided Student's t‐test (**P < 0.01; ****P < 0.0001, n = 4). Given are mean ± the standard error of the mean. * indicates an unspecific band. Surface and lysate samples are separated by a marker lane.
-
BWt neurons were infected with NrCAM shRNA‐containing lentiviruses (shRNA1 or shRNA2; 1:1,000), or a scrambled control construct (1:1,000). At DIV4, the cells were lysed and the conditioned media were collected.
-
CCell viabilities after infection with the respective viruses (scr., sh1 and sh2) were compared to a positive (no toxic effect) and negative control (high toxicity) with Cell Counting Kit 8 (Sigma). The luminescence signals were measured with a microplate reader. Both shRNAs did not show higher toxicity than the scrambled control virus. Pos. = positive survival control (no treatment); neg. = negative survival control (cells treated with NaOH). One‐way ANOVA with post hoc Dunnett's test (n.s P > 0.05; **P < 0.01; ****P < 0.0001, n = 8). P‐values were calculated by comparing the respective treatments to the treatment with the scr. shRNA. Shown are mean and SEM. Representative Western blots are shown.
-
DqPCRs for the ADAM10 and NrCAM mRNAs from wt neurons treated with GI254023x or solvent for 48 h (preparation and treatment of the cells were performed like in Fig 3B). Two‐sided Student's t‐test (n = 5–6). Given are mean ± the standard error of the mean.
Source data are available online for this figure.
