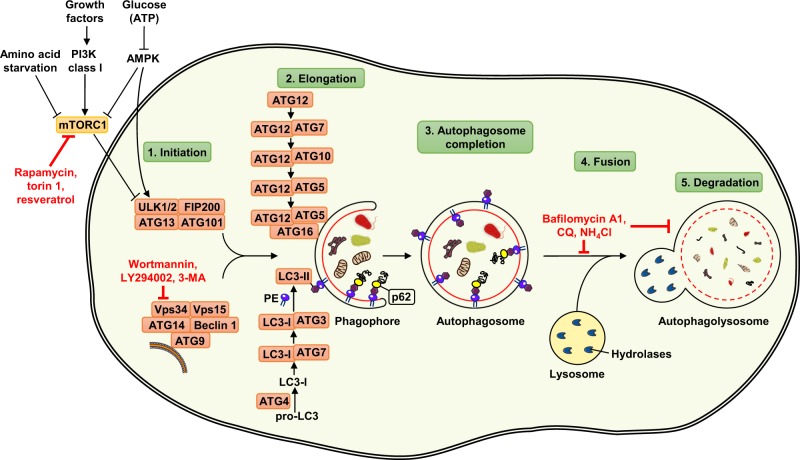
Fig. 1.
Mechanism of autophagy and its regulation in mammalian cells. The autophagy machinery is regulated by different functional groups of ATG proteins. By integrating several upstream signals, autophagy is initiated by the activation of ULK kinase complex, consisting of proteins ULK1/2, ATG13, ATG101, and FIP200. The autophagic process is inhibited in the presence of growth factors through the action of the class I PI3K-mTORC1 axis, whereas autophagy is induced by amino acid starvation or lack of energy through the AMPK pathway. The pharmacological inhibitors rapamycin, torin 1, and resveratrol act as mTORC1 inhibitors and therefore induce autophagy. Nucleation of the autophagosomal membrane is also regulated by the class III PI3K complex, consisting of proteins Beclin 1, ATG14, ATG9, Vps34, and Vps15. Wortmannin, LY294002, and 3-MA are commonly used autophagy inhibitors, acting as blockers of class III PI3K (known as Vps34) enzymatic activity. ATG proteins that are involved in the elongation process belong to two conjugation systems: ATG12 covalently binds to ATG5 with the assistance of ATG7 and ATG10 enzymes, forming a complex through interaction with ATG16. A second conjugation system leads to conjugation of LC3-I with PE. The lipidated LC3-II exists as a part of the autophagosomal membrane, and allows p62 to target ubiquitinated cargo to the expanding autophagosomal membrane by binding to LC3-II. The completed autophagosome fuses with the lysosome, and the autophagolysosomal contents are degraded by lysosomal hydrolytic enzymes. Chemical agents, such as CQ, bafilomycin A1, and ammonium chloride, can inhibit degradation of engulfed cytoplasmic material as well as the fusion between autophagosomes and lysosomes