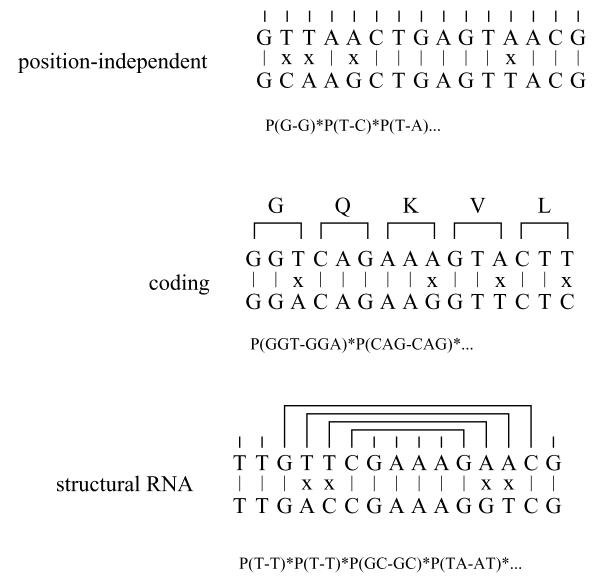
Figure 1.
Three pairwise alignments of identical composition with identical number of base substitutions can be classified by distinctive patterns of mutation caused by different selective constrains: the position-independent null hypothesis (top), a coding region (middle), or a structural RNA (bottom). We indicate how each alignment is scored according to the model that best fits the pattern of mutations: one position at the time for OTH, one codon at the time for COD (integrated over all six possible frames), and as a combination of base-paired positions and single positions for RNA (integrated over all possible secondary structures).