Short abstract
Objectives
Acute leukemia (AL) is a highly heterogeneous malignant disease caused by hematopoietic cell abnormalities. Our study investigated the potential for immunophenotyping of leukemic cells via flow cytometry and the clinical usefulness of this approach in treatment of AL.
Methods
Bone marrow (BM) specimens were collected to detect antigen expression on hematopoietic cells in pre-treatment samples from patients with AL. In addition, fraction survival curves were calculated using the Kaplan-Meier method to explore the effect of markers on prognosis in AL.
Results
Expression levels of immunophenotypic markers in patients with acute lymphoblastic leukemia (ALL) were significantly different from those in patients with acute myeloid leukemia (AML). In addition, there was a potential association between the surface marker, cluster of differentiation 2 (CD2), and fraction survival in AML. However, no similar result was found in ALL. Moreover, genetic tests showed greater positive variation of the break point cluster-Abelson tyrosine kinase (BCR-ABL) fusion gene in samples from patients with ALL than in samples from patients with AML.
Conclusions
We have shown a rapid and effective flow cytometry method that enables the identification of immunophenotype in AL. Moreover, CD2 may constitute a predictive marker for prognosis in patients with AML.
Keywords: Acute leukemia, myeloid leukemia, acute lymphoblastic leukemia, flow cytometry, immunophenotype, bone marrow, CD2, break point cluster-Abelson tyrosine kinase fusion gene
Introduction
Acute leukemia (AL) is a highly heterogeneous malignant disease caused by hematopoietic cell abnormalities.1,2 Based on the morphologic, immunologic, cytogenetic, molecular, and clinical features, AL is divided into two major types: acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL).2 Moreover, AML is further divided into eight subtypes (M0–M7) based on the cell type of origin and its degree of maturity,3 while ALL is classified into three subtypes (L1, L2, and L3).4
Cytogenetic analysis using World Health Organization (WHO) classification is the gold standard and predominant method for assessment of AL; however, this approach is often not available for preliminary diagnosis in clinical practice.5,6 Furthermore, reliable recognition of different types and subtypes of AL may require additional tests, such as fluorescent in situ analysis, because abnormalities may be not always detected by routine cytogenetic analysis.
Flow cytometry immunophenotyping (FCI) is a powerful technology that can be used to identify cell membrane antigens.7 The identification of antigens on leukemic cells is helpful to guide administration of specific treatments for patients.8 It is also helpful to estimate the prognosis of AL patients and search for applicable markers to detect minimal residual disease.
Immunophenotyping could enable the detection of leukemic stem cells, both at time of diagnosis and follow-up; moreover, it may be useful to assess disease during emergence and development.9,10 A recent study reported that peripheral blood minimal residual disease may replace bone marrow (BM) minimal residual disease with respect to immunophenotypic biomarkers to predict AML relapse.11
In this study, we aimed to determine whether ALL and AML exhibited different immunophenotypes and to explore the clinical usefulness of these differences as ancillary tools in patients with AL.
Materials and methods
Subjects and ethical approval
Immunophenotyping data were obtained from patients with AL who were treated at Ningbo First Hospital during the period from January 2001 to August 2013. Immunophenotyping was performed using BM specimens obtained before treatment. The diagnosis of AL patients was made by using the strict classifications of the French American British (FAB) co-operative group and the WHO.3,9 Exclusion criteria were a history of hematopoietic cell transplantation, uncontrolled active infection, treatment with cytokines within the previous 4 weeks, New York Heart Association (NYHA) grade IV heart failure, other malignancy not in remission, and/or severe psychiatric disorder. This study was approved by the Human Research Ethics Committees of Ningbo First Hospital and Ningbo University. All patients provided written informed consent for BM collection and immunophenotyping research.
Flow cytometric immunophenotyping
In a separate microcentrifuge tube for each sample, heparin-anticoagulation BM (100 µL) was stained with fluorochrome-conjugated antigen-specific monoclonal antibodies (20 µL). The mixed sample was incubated for 15 minutes at room temperature, and was shielded from light. Next, hemolytic agent was added to the tube and the suspension was mixed; the sample was incubated for 10 minutes at room temperature, and was shielded from light. Phosphate-buffered saline (PBS) was then added to the tube and the sample was centrifuged at 1500 × g for 5 minutes. Thereafter, the supernatant was discarded and the sample was used for flow cytometry analysis. For staining of cytoplasmic antigens, cells were permeabilized and fixed with a cell fixation and permeabilization kit (FIX & PERM, Kaumberg, Austria).12 After cell preparations had undergone a final washing procedure, the cells were resuspended in PBS for flow cytometry analysis.
A comprehensive antibody with four-color direct immunofluorescent labeling was used for immunophenotyping with a BD FACSCanto II System (BD Biosciences, San Diego, CA, USA). The blast population was gated on the basis of light scattering properties. Unstained cells in each tube were used as negative controls to set quadrant gates. The percentage of antigen-positive cells (cut off > 20%) was calculated using the quadrant statistics. Expression of one or more markers on more than 20% of the blast cell population was considered a positive result.12 The reagent system was a panel of monoclonal antibodies (Immunotech, Marseille, France) that was used to detect clusters of differentiation (CD) and other antigens (myeloperoxidase, MPO; human leukocyte antigen - DR isotype, HLA-DR); these included myeloid markers (MPO, CD13, CD14, CD15, CD33, CD117, and CD11b), T-cell lineage markers (CD2, CD3, cyCD3, CD5, and CD7), B-cell lineage markers (CD10, CD19, and CD20), megakaryocytic lineage markers (CD41 and CD61) and other markers (CD34, CD38, HLA-DR, CD71, CD9, and CD56).
Survival analysis and fusion gene tests
Fraction survival was measured from the date of diagnosis of AL. Patients known to be alive at the time of the last follow-up were censored on the last date of contact. The fraction survival curves were calculated with the Kaplan-Meier method and were analyzed using the log-rank test. The break point cluster-Abelson tyrosine kinase (BCR-ABL) fusion gene was used to evaluate the status of BM samples with a leukemia fluorescence quantitative polymerase chain reaction (qPCR) diagnostic kit (Yuanqi, Shanghai, China).
Statistical analysis
Statistical tests were conducted using the Statistical Program for Social Sciences (SPSS) software 17.0 (SPSS Inc., Chicago, IL, USA). Values of markers that deviated from normality were corrected via logarithmic transformation. A nonparametric approach was used to analyze data that could not be normalized. Student’s t-test and one-way analysis of variance were used to determine statistical significance. A two-sided P < 0.05 was considered to be significant.
Results
Clinicopathologic features
The diagnostic criteria of the AL patients in this study comprised morphological examination and cytochemical staining, as well as the percentage of blast cells. A rate of blast cells > 20% of the marrow karyotype population was considered positive according to the WHO classification criteria. Our study recruited a total of 143 samples from patients with ALL and 470 samples from patients with AML (613 total patients with AL). The patients with AML included 11 with AML-M1, 52 with AML-M2, 106 with AML-M3, 125 with AML-M4, 151 with AML-M5, 11 with AML-M6, and 14 with other AML (Table 1). The male-to-female ratios of patients with AML and ALL were 1.03:1 and 1:1, respectively. The mean values and standard deviations of patients’ ages were 48.23 ± 17.75 years (AML) and 37.52 ± 17.81 years (ALL), as shown in Table 1.
Table 1.
Significant immunophenotype differences between M3 and all other AML subtypes.
| P value between M3 and | M1 (n = 11) | M2 (n = 52) | M4 (n = 125) | M5 (n = 151) | M6 (n = 11) |
|---|---|---|---|---|---|
| CD13 | 0.017 | 0.004 | 5.94E-06 | 1.43E-06 | 0.047 |
| CD14 | 0.386 | 0.802 | 0.030 | 0.009 | 0.002 |
| CD15 | 0.021 | 0.863 | 0.343 | 0.837 | 0.383 |
| CD33 | 0.006 | 1.89E-04 | 1.08E-05 | 1.17E-09 | 1.86E-04 |
| CD117 | 0.328 | 0.004 | 4.94E-09 | 0.007 | 0.268 |
| CD11b | 0.913 | 0.457 | 0.011 | 0.008 | 0.103 |
| MPO | 0.312 | 0.078 | 0.791 | 0.422 | 0.861 |
| CD2 | 0.273 | 0.021 | 1.77E-04 | 0.063 | 0.413 |
| CD3 | 0.138 | 0.461 | 0.637 | 0.868 | 0.184 |
| cyCD3 | 0.529 | 0.064 | 0.089 | 0.280 | 0.487 |
| CD5 | 0.141 | 0.069 | 0.004 | 4.99E-04 | 0.014 |
| CD7 | 0.056 | 3.69E-05 | 4.90E-10 | 4.73E-09 | 3.32E-04 |
| CD10 | 0.225 | 0.041 | 0.128 | 0.032 | 0.022 |
| CD19 | 0.609 | 1.99E-04 | 0.007 | 0.222 | 0.017 |
| CD20 | 0.962 | 0.404 | 0.671 | 0.217 | 0.022 |
| CD41 | 0.421 | 0.021 | 0.027 | 4.90E-04 | 0.003 |
| CD61 | 0.042 | 0.505 | 0.077 | 0.040 | – |
| CD34 | 0.256 | 3.87E-11 | 8.05E-18 | 1.68E-14 | 3.41E-04 |
| CD38 | 0.265 | 6.08E-09 | 2.14E-14 | 6.03E-10 | 0.678 |
| HLA-DR | 0.069 | 6.54E-10 | 3.07E-18 | 5.29E-20 | 2.28E-06 |
| CD71 | 0.431 | 0.154 | 0.753 | 0.766 | 0.010 |
| CD9 | 5.06E-05 | 3.85E-16 | 1.02E-24 | 5.83E-18 | 2.55E-05 |
| CD56 | 0.923 | 0.261 | 0.165 | 0.950 | 0.452 |
AML: acute myeloid leukemia; CD: clusters of differentiation; HLA-DR: human leukocyte antigen-DR isotype.
Immunophenotypic findings
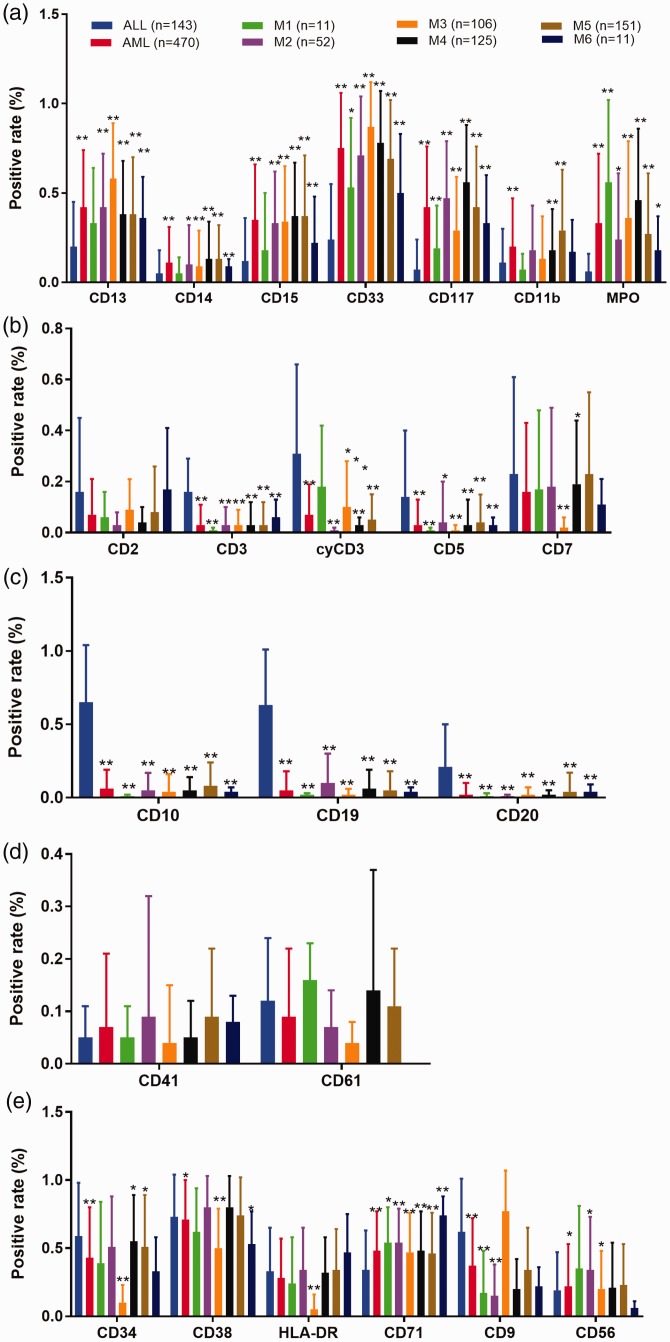
As shown in Figure 1, we identified significant differences in immunophenotypic biomarkers between patients with ALL and those with AML. Regarding myeloid markers (Figure 1a), expression levels were generally higher in patients with AML than in those with ALL. In subtype analysis, most subtypes showed significantly different levels of myeloid markers among the various subtypes of ALL and AML; the differences between M1 and ALL were moderate. Among all myeloid markers, CD11b showed no significant difference, compared with other biomarkers. CD33 was the most frequently expressed antigen in samples from both patient groups (ALL and AML).
Figure 1.
Significant differences in immunophenotypic biomarkers between acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). (a) Myeloid markers. (b) T-cell lineage markers. (c) B-cell lineage markers. (d) Megakaryocytic lineage markers. (e) Other markers. *P < 0.05; **P < 0.01.
Regarding T-cell lineage markers (Figure 1b), expression levels were generally higher in patients with ALL than in patients with AML. The expression levels of CD3, cyCD3, and CD5 were significantly different between patients with ALL and those with AML; moreover, they were significantly different among the various subtypes of AML. Expression levels of B-cell lineage markers were generally higher in patients with ALL than in those with AML (Figure 1c). The expression levels of CD10, CD19, and CD20 were significantly different between patients with ALL and those with AML, as well as among the various subtypes of AML. The levels of megakaryocytic lineage markers were not statistically different between patients with ALL and those with AML (Figure 1d). CD41 levels were significantly different between ALL and M5-6 AML.
Regarding the other markers (Figure 1e), there were differing levels between patients with ALL and those with AML. The expression levels of CD71 were significantly different between patients with ALL and those with AML, as well as among the various subtypes of AML. CD9 expression was significantly higher in patients with ALL than in those with AML. In addition, there were differences between patients with ALL and those with subtypes of AML in CD34, CD38, and CD56. Among these three markers, CD38 was the most highly expressed antigen in patients with ALL and in those with most subtypes of AML.
Considering that M3 is a unique subtype of AML that does not require curative BM transplantation, we further compared the levels of antigens among M3 and other subtypes of AML. There were significant differences in the expression of antigens among AML subtypes, particularly with respect to CD33, CD7, CD34, CD38, HLA-DR, and CD9, as shown in Table 1. However, the expression levels of biomarkers showed no significant differences between M1 and M3.
Immunophenotypic biomarkers may be associated with survival in AML
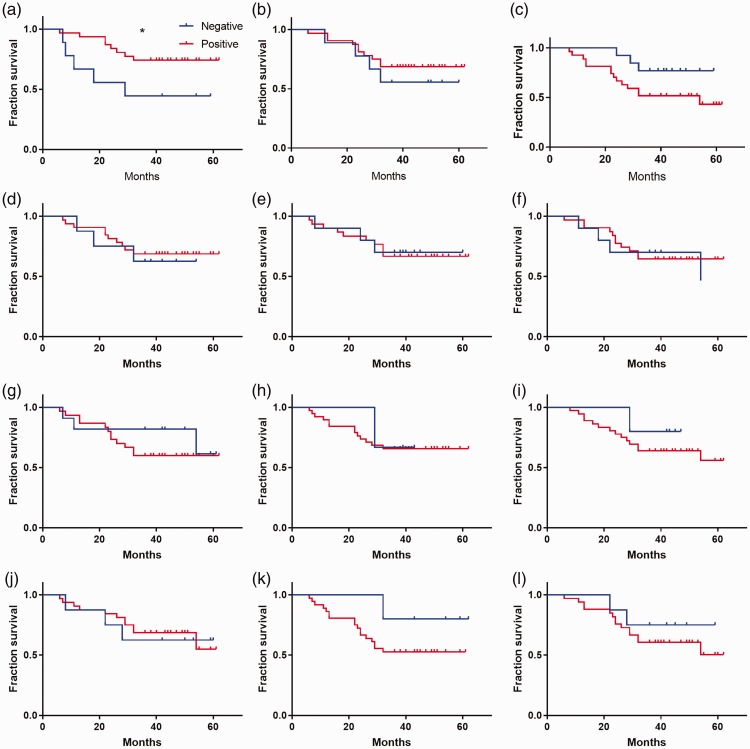
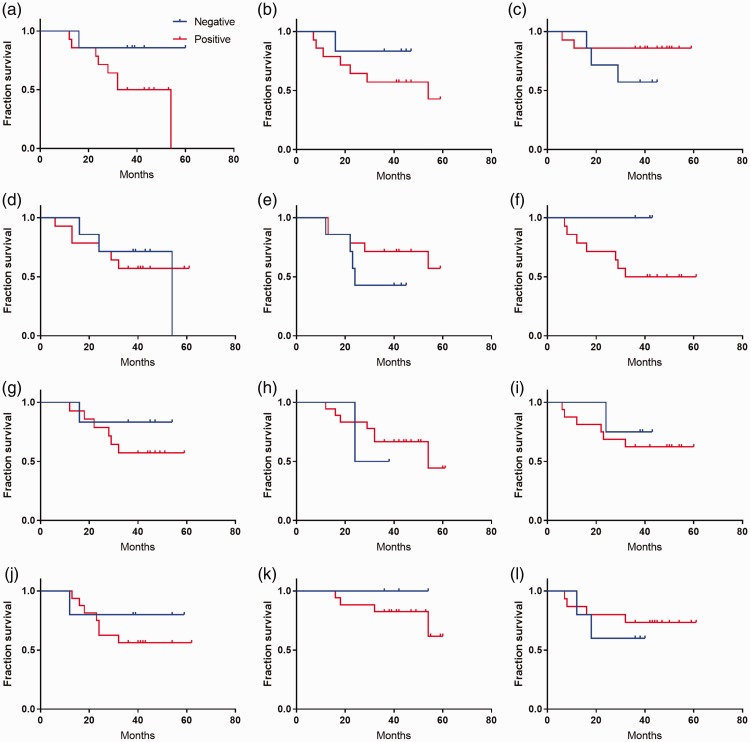
Because there were significant differences in the expression levels of immunophenotypic biomarkers between patients with ALL and those with AML, we assessed whether these immunophenotypic biomarkers could serve as prognostic markers for AL. Our preliminary results showed an association between the level of CD2 and fraction survival in patients with AML (Figure 2a, log-rank *P < 0.05). However, no association was found between fraction survival and any of the immunophenotypic biomarkers in patients with ALL (Figure 3).
Figure 2.
Association between fraction survival and immunophenotypic biomarkers in patients with acute myeloid leukemia (log-rank, *P < 0.05). (a) CD2. (b) CD5. (c) CD7. (d) CD3. (e) CD56. (f) CD19. (g) CD20. (h) CD13. ((i) CD117. (j) CD14. (k) HLA-DR. (l) CD10.
Figure 3.
Lack of association between fraction survival and classification of immunophenotypic biomarkers in patients with acute lymphoblastic leukemia (log-rank, P > 0.05). (a) CD2. (b) CD5. (c) CD7. (d) CD3. (e) CD56. (f) CD19. (g) CD20. (h) CD13. ((i) CD117. (j) CD14. (k) HLA-DR. (l) CD10.
Genetic findings
As shown in Table 2, we found that almost all patients with AML were BCR-ABL p190- and p210-negative, whereas some patients with ALL were BCR-ABL p190- and p210-positive. Promyelocytic leukemia protein-positive samples were detected only in patients with AML-M3.
Table 2.
Chromosome and molecular genetics results in the ALL and AML cases.
| BCR p190 (+) | BCR p190 (−) | BCR p210 (+) | BCR p210 (−) | PML (+) | PML (−) | |
|---|---|---|---|---|---|---|
| ALL (n = 143) | 8 | 17 | 3 | 29 | 0 | 11 |
| M1 (n = 11) | 0 | 1 | 0 | 3 | 0 | 0 |
| M2 (n = 52) | 0 | 2 | 0 | 3 | 0 | 5 |
| M3 (n = 106) | 0 | 2 | 0 | 7 | 25 | 8 |
| M4 (n = 125) | 0 | 7 | 0 | 28 | 0 | 13 |
| M5 (n = 151) | 0 | 10 | 0 | 25 | 0 | 13 |
| M6 (n = 11) | 0 | 0 | 0 | 1 | 0 | 0 |
| M7 (n = 1) | 1 | 0 | 0 | 1 | 0 | 1 |
ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; BCR: break point cluster; PML: promyelocytic leukemia protein.
Discussion
AL is a highly heterogeneous group of malignancies that exhibit different clinical features and prognoses; these require specific therapeutic regimens.13 Timely and accurate diagnosis is critical for the treatment of AL.14 FCI has been used to detect specific surface and cytoplasmic antigens of leukemic cells to determine the sources and differentiation stages of leukemia with monoclonal antibodies.14,15 The analysis of a large number of cells can be completed quickly by flow cytometry, which improves the accuracy of leukemia diagnosis.6,11 In addition, immunophenotyping may reflect biological characteristics that the FAB and WHO classification systems do not discuss, such as a double phenotype and the stage of ALL. These biological characteristics may affect the management of treatment protocol in patients with either AML or ALL, as well as the estimation of prognosis in patients with ALL.
Malignant cells often have aberrant phenotypes that may help to distinguish them from normal immature cells. One of these aberrations is the presence of phenotypes that are not typically present on that particular cell lineage.16 For instance, although CD56 is a lymphoid marker, it could be detected in both ALL and AML.17 Moreover, patients with AML who were CD56-positive exhibited a worse prognosis.7,18 Significantly different levels of antigens (CD2, CD5, CD7, sCD3, CD19, CD20, sCD13, CD33, CD117, CD15, CD14, CD34, CD10, CD71, CD9, CD41, MPO, and cCD3) were observed between patients with AML and those with ALL (Figure 1). These antigens comprised markers of myeloid cells, B lymphocytes, T lymphocytes, and megakaryocytes, all of which are useful in the classification of leukemia. However, there were no significant differences between patients with AML and those with ALL in the levels of CD56, HLA-DR, CD38, CD61, and cCD13. These results indicated that specific antigens may indeed act as potential molecular markers for the classification of AL subtypes.
HLA-DR reportedly is expressed in both AML and ALL.19 Furthermore, it has shown significantly different levels among patients with standard-risk, medium-risk, and high-risk status in ALL.16 Thus, HLA-DR might be useful in risk evaluation for patients with AML and those with ALL.3 Our results indicated that CD2 level may be associated with fraction survival in patients with AML (Figure 2, *P < 0.05). Although there was a related trend in the association analysis between fraction survival and CD7 level in patients with AML, there was no significant impact of other aberrant markers on prognosis (Figure 2). Thus, these associations may provide hints to identify useful disease markers and prognostic factors for predicting remission in patients with AML.3
In this study, we found different outcomes based on fusion gene analysis in patients with ALL, those with AML, and the subtypes of both AML and ALL. Previous studies revealed that patients AL who exhibit BCR-ABL mutations seemed to have a worse prognosis than those without these mutations,3 demonstrating that the detection of fusion genes was important for the diagnosis, classification, and prognosis of AL. In the present study, most AL subtypes showed different fusion gene statuses (Table 2). These results provided new evidence to support the findings of the previous study.3 Unfortunately, we did not perform analysis of cytogenetic-immunophenotype correlations due to the small sample size in this study. A larger sample size with corresponding cytogenetic information is needed to explore this association in future studies.
In conclusion, our results showed that immunophenotypes varied significantly between patients with AML and those with ALL, including among their various subtypes; this indicated that FCI may provide a clinically useful, rapid, and effective method to aid in diagnosis in patients with AL, as well as to assess treatment progress and predict prognosis.
Declaration of conflicting interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the K. C. Wong Magna Fund in Ningbo University. The sponsors had no role in the study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
References
- 1.Hsiao PC, Liu MC, Chen LM, et al. Promoter methylation of p16 and EDNRB gene in leukemia patients in Taiwan. Chin J Physiol 2008; 51: 27–31. [PubMed] [Google Scholar]
- 2.Petrov I, Suntsova M, Mutorova O, et al. Molecular pathway activation features of pediatric acute myeloid leukemia (AML) and acute lymphoblast leukemia (ALL) cells. Aging (Albany NY) 2016; 8: 2936–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol 1976; 33: 451–458. [DOI] [PubMed] [Google Scholar]
- 4.Davey FR, Castella A, Lauenstein K, et al. Prognostic significance of the revised French-American-British classification for acute lymphocytic leukaemia. Clin Lab Haematol 1983; 5: 343–351. [DOI] [PubMed] [Google Scholar]
- 5.Wang XB, Zheng JE, Gu JX, et al. [Correlation of immunophenotype to cytogenetics and clinical features of adult acute myeloid leukemia]. Ai Zheng 2005; 24: 667–671. [PubMed] [Google Scholar]
- 6.Iriyama N, Asou N, Miyazaki Y, et al. Normal karyotype acute myeloid leukemia with the CD7+ CD15+ CD34+ HLA-DR + immunophenotype is a clinically distinct entity with a favorable outcome. Ann Hematol 2014; 93: 957–963. [DOI] [PubMed] [Google Scholar]
- 7.Xu B, Hu C, Miao XD, et al. [Immunophenotyping of 106 adult patients with acute leukemia by flow cytometry]. Di Yi Jun Yi Da Xue Xue Bao 2003; 23: 1043–1046. [PubMed] [Google Scholar]
- 8.Khoury H, Dalal BI, Nevill TJ, et al. Acute myelogenous leukemia with t(8; 21)–identification of a specific immunophenotype. Leuk Lymphoma 2003; 44: 1713–1718. [DOI] [PubMed] [Google Scholar]
- 9.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009; 114: 937–951. [DOI] [PubMed] [Google Scholar]
- 10.Zeijlemaker W, Kelder A, Oussoren-Brockhoff YJ, et al. A simple one-tube assay for immunophenotypical quantification of leukemic stem cells in acute myeloid leukemia. Leukemia 2016; 30: 439–446. [DOI] [PubMed] [Google Scholar]
- 11.Zeijlemaker W, Kelder A, Oussoren-Brockhoff YJ, et al. Peripheral blood minimal residual disease may replace bone marrow minimal residual disease as an immunophenotypic biomarker for impending relapse in acute myeloid leukemia. Leukemia 2016; 30: 708–715. [DOI] [PubMed] [Google Scholar]
- 12.Ratei R, Schabath R, Karawajew L, et al. Lineage classification of childhood acute lymphoblastic leukemia according to the EGIL recommendations: results of the ALL-BFM 2000 trial. Klin Padiatr 2013; 225: S34–S39. [DOI] [PubMed] [Google Scholar]
- 13.Snijder B, Vladimer GI, Krall N, et al. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. Lancet Haematol 2017; 4: e595–e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweet K, Lancet J. State of the art update and next questions: acute myeloid leukemia. Clin Lymphoma Myeloma Leuk 2017; 17: 703–709. [DOI] [PubMed] [Google Scholar]
- 15.Han X, Medeiros LJ, Zhang YH, et al. High expression of human homologue of murine double minute 4 and the short splicing variant, HDM4-S, in bone marrow in patients with acute myeloid leukemia or myelodysplastic syndrome. Clin Lymphoma Myeloma Leuk 2016; 16: S30–S38. [DOI] [PubMed] [Google Scholar]
- 16.Al-Mawali A, Gillis D, Hissaria P, et al. Incidence, sensitivity, and specificity of leukemia-associated phenotypes in acute myeloid leukemia using specific five-color multiparameter flow cytometry. Am J Clin Pathol 2008; 129: 934–945. [DOI] [PubMed] [Google Scholar]
- 17.Abdulateef NA, Ismail MM, Aljedani H. Clinical significance of co-expression of aberrant antigens in acute leukemia: a retrospective cohort study in Makah Al Mukaramah, Saudi Arabia. Asian Pac J Cancer Prev 2014; 15: 221–227. [DOI] [PubMed] [Google Scholar]
- 18.Ossenkoppele GJ, van de Loosdrecht AA, Schuurhuis GJ. Review of the relevance of aberrant antigen expression by flow cytometry in myeloid neoplasms. Br J Haematol 2011; 153: 421–436. [DOI] [PubMed] [Google Scholar]
- 19.Hur M, Chang YH, Lee DS, et al. Immunophenotypic and cytogenetic changes in acute leukaemia at relapse. Clin Lab Haematol 2001; 23: 173–179. [DOI] [PubMed] [Google Scholar]