Short abstract
Objective
The aim of this study was to identify important pathways regulated by a set of long non-coding RNAs (lncRNAs) in oral squamous cell carcinoma (OSCC).
Methods
A lncRNA-mediated competitive endogenous RNA network (LMCN) was constructed using information on microRNA (miRNA)–mRNA interactions and lncRNA–miRNA intersections from the E-GEOD-37991 transcription profiling data in the ArrayExpress database. A random walk with restart ranking algorithm was then applied to evaluate the influences of protein-coding genes regulated by competitive lncRNAs. Pathway enrichment scores were calculated based on the propagation scores of protein-coding genes. Finally, permutation tests were used to estimate the significance of the pathways.
Results
We obtained lncRNA–mRNA interactions based on miRNAs common to both miRNA–mRNA interactions and lncRNA–miRNA intersections, and used interactions with a z-score > 0.7 to construct a LMCN. Ten lncRNAs were identified as source nodes in the LMCN, and nine pathways with enrichment scores >0.8, including ‘Cell cycle’, ‘Endocytosis’, and ‘Pathways in cancer’, were significantly enriched by these source nodes.
Conclusions
Nine significant pathways regulated by a set of competitive lncRNAs were identified in OSCC, which may play important roles in the development of OSCC via the cell cycle and endocytosis.
Keywords: lncRNA, oral squamous cell carcinoma, pathway, cell cycle, endocytosis, network
Introduction
Oral cancer may occur in the lips, floor of the mouth, upper and lower alveolar ridges, buccal mucosa, hard palate, sublingual region, retromolar trigone, and anterior two-thirds of the tongue.1 Oral cancer is the third most common carcinoma in developing countries and the sixth most common carcinoma worldwide,2 with oral squamous cell carcinoma (OSCC) accounting for more than 95% of all malignancies in the oral cavity.3 Despite significant progress in the diagnosis and clinical treatment of OSCC in recent decades, the overall survival rate has not improved significantly.4
Long non-coding RNAs (lncRNAs) are a class of non-protein coding RNAs of more than 200 nucleotides in length, which have been discovered in a wide range of biological processes to date.5 Many of these lncRNAs have been shown to be uniquely expressed in specific cancer types or differentiated tissues, and increasing evidence suggests that lncRNAs perform key roles in promoting cancer development and progression.6 LncRNA-UCA1 has been proposed to play an oncogenetic role in OSCC through regulating the Wnt/b-catenin signaling pathway,7 while lncRNA-MALAT1 was found to promote tumor metastasis and growth through inducing epithelial-mesenchymal transition in OSCC.8
Although dysregulation of lncRNAs has been shown in OSCC, lncRNA profiles in this type of cancer remain largely unknown. In this study, we applied a novel method, LncRNAs2Pathways,9 to identify important lncRNA-related pathways in OSCC. LncRNAs2Pathways identifies pathways influenced by the set of lncRNAs of interest based on a global network propagation algorithm.9 This method has been successfully applied in glioma, and in pancreatic and prostate cancers, with most of the identified pathways proposed to be related to the progression of prostate cancer.9
The aim of this study was to characterize key pathways regulated by a set of competitive lncRNAs in OSCC. We collected microRNA (miRNA)–mRNA interactions and lncRNA–miRNA intersections from the starBase v2.0 database, and transcription profiling data from the ArrayExpress database and processed them using the LncRNAs2Pathways method to identify significant pathways likely to be regulated by a set of competitive lncRNAs.
Materials and methods
Data
Transcription profiling data in E-GEOD-3799110, which includes transcriptional filing data for tumor and paired non-tumor samples, were downloaded from the ArrayExpress database. Control (n = 40) and OSCC data (n = 40) were collected. According to the probe ID and gene symbol in the annotation file, the probe ID was changed to its gene symbol using the feature function Filter.11 A total of 18,631 genes were finally obtained.
A total of 10,212 pairs of lncRNA–miRNA intersections and 423,975 pairs of miRNA–mRNA interactions were downloaded from the starBase v2.0 database.
The 18,631 genes with transcription data were mapped to the lncRNA–miRNA intersections and miRNA–mRNA interactions, and mRNAs (n = 9125) and lncRNAs (n = 10) common to both were collected as new profiling data (n = 9135). Interactions containing genes from the new profiling data were then collected. A total of 221 pairs of lncRNA–miRNA intersections and 293,185 pairs of miRNA–mRNA interactions were finally obtained.
Construction of competitive endogenous RNA (ceRNA) network
We evaluated the significance of the miRNAs common to both mRNA–miRNA interactions and lncRNA–miRNA interactions using a hypergeometric test, as follows:12
| (1) |
The P-values were subjected to false discovery rate correction. An adjusted P < 0.05 was used as the threshold. Finally, 10 lncRNAs, 6206 mRNAs, and 10,772 interactions were obtained.
We characterized the lncRNA–mRNA interactions by calculating Pearson’s correlation coeffcients according to the expression of the competing lncRNA–mRNAs interactions. Correlation coefficients were then converted to the z-statistic according to Fisher’s z transform method,13 as follows:
| (2) |
where r is Pearson’s correlation coefficient. LncRNA–mRNA interactions with z > 0.7 were considered as co-expression networks and used to construct a lncRNA-mediated ceRNA (LMCN) network, including 10 lncRNAs and 481 mRNAs. The lncRNAs in the LMCN network were considered as source nodes.
Evaluation of influence of protein-coding genes regulated by competitive lncRNAs
Protein-coding genes are more likely to be regulated if they are located close to the source nodes. We evaluated the influence of protein-coding genes regulated by competitive lncRNAs using the random walk with restart ranking algorithm,14 as follows:
| (3) |
where r indicates the restart probability of the walk in every time step at the source node. In this study, r was set at 0.7.
The probabilities of protein-coding genes were then normalized and defined as propagation scores. All the protein-coding genes were ranked in the gene list L, based on their propagation scores.
Calculating pathway enrichment scores (ES) to evaluate biological pathways
Pathway ES were calculated based on propagation scores of protein-coding genes. Human-related pathways were downloaded from the Kyoto Encyclopedia of Genes and Genomes database. Pathways with > 500 or < 15 protein-coding genes were deleted.
Protein-coding genes in pathways were mapped to the gene list L. The mapped pathway was more likely to be influenced by the combinatorial effects of the competitive lncRNAs if the genes in that pathway occurred near the top of the list L. We identified ES according to the method proposed by Han et al.,9 as follows:
| (4) |
| (5) |
| (6) |
| (7) |
where P was set at 1.
Pathway significance analysis
We estimated the significance of the pathway ES using permutation tests.9 After 1000 permutation tests, the P-value of the observed ES was calculated by comparison with the set of scores in ESNULL, shown as P-value = M/N, where N indicates the number of permutation tests and M indicates the number of ESNULL values that were greater than the observed ES. The P-value was then adjusted by the false discovery rate,15 and pathways with adjusted P < 0.05 were selected as target pathways regulated by the set of competitive lncRNAs.
No animal or human tissue or cells were directly involved in this study. The transcription profiling data were obtained from the ArrayExpress database and ethical permission was therefore unnecessary.
Results
Construction of LMCN network
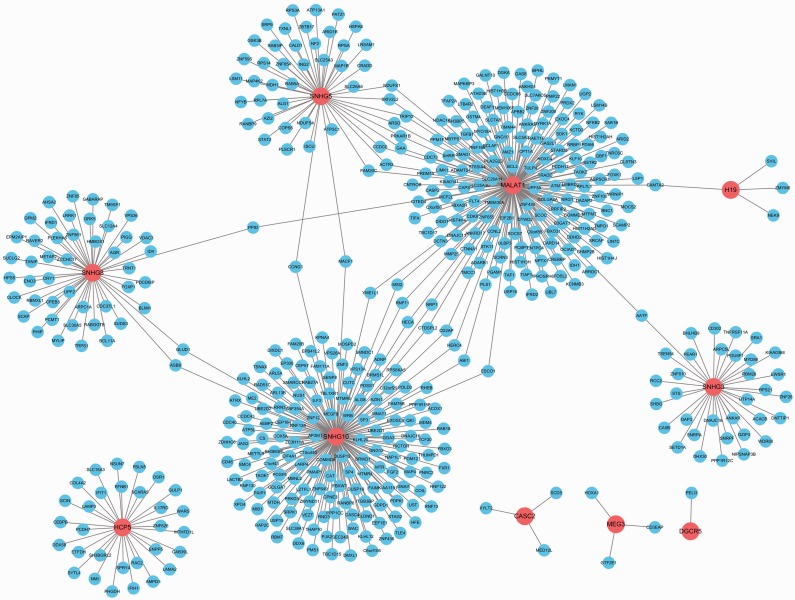
We identified lncRNA–mRNA interactions based on common miRNAs between lncRNA–miRNA intersections and miRNA–mRNA interactions, and used interactions with a z-score > 0.7 to construct the LMCN network. The network included 10 lncRNAs and 481 mRNAs (Figure 1).
Figure 1.
Long non-coding RNA (lncRNA)-mediated competitive endogenous RNA network. The network included 10 lncRNAs and 481 mRNAs. Competitive lncRNAs were considered as source nodes, including MALAT1, SNHG10, SNHG5, SNHG8, HCP5, SNHG3, H19, MEG3, DGCR5, and CASC2
Evaluation of influence of protein-coding genes regulated by competitive lncRNAs
We evaluated the influence (propagation scores) of protein-coding genes regulated by competitive lncRNAs and ranked the protein-coding genes in the gene list L, according to their propagation scores. We considered that the greater the probability of a protein-coding gene being located closer to the source nodes, the more this gene was likely to be influenced by the source nodes.
Identification of significant pathways
Based on pathway ES, we performed 1000 permutation tests to estimate the significance of the pathways. Nine significant pathways with ES values >0.8 were finally identified, including ‘Cell cycle’, ‘Endocytosis’, ‘Pathways in cancer’, ‘Chemokine signaling pathway’, ‘Axon guidance’, ‘Spliceosome’, ‘Focal adhesion’, ‘MAPK signaling pathway’, and ‘Regulation of actin cytoskeleton’ (Table 1).
Table 1.
Pathways regulated by the set of competitive long non-coding RNAs
| Pathway | Enrichment score | P-value | Adj. P-value |
|---|---|---|---|
| Cell cycle | 0.9 | <0.01 | <0.01 |
| Endocytosis | 0.89 | <0.01 | <0.01 |
| Pathways in cancer | 0.89 | <0.01 | <0.01 |
| Chemokine signaling pathway | 0.85 | <0.01 | <0.01 |
| Axon guidance | 0.85 | <0.01 | <0.01 |
| Spliceosome | 0.84 | <0.01 | <0.01 |
| Focal adhesion | 0.83 | <0.01 | <0.01 |
| MAPK signaling pathway | 0.83 | <0.01 | <0.01 |
| Regulation of actin cytoskeleton | 0.81 | <0.01 | <0.01 |
Adj. P-value: P-value adjusted by false discovery rate
Discussion
In this study, we applied the LncRNAs2Pathways method to identify important pathways regulated by a set of lncRNAs of interest. Nine pathways were identified, of which the top three pathways were ‘Cell cycle pathway’, ‘Endocytosis pathway’, and ‘Pathways in cancer’.
‘Cell cycle pathway’ plays a vital role in cell differentiation, angiogenesis, and cell proliferation and survival. Cell cycle progression of mitotic cells is accomplished via a sequence of events, including DNA replication and mitosis, with temporal gaps known as G1 and G2 phases. Several genes in this pathway have been reported to be closely related to cancers, including the genes encoding cyclin-dependent kinases, p53, and retinoblastoma protein (Rb). Cyclin-dependent kinases are critical regulatory enzymes affecting all cell cycle transitions.16 p53 is a tumor suppressor protein that has been widely reported in many kinds of human cancers,17,18 and p53 mutation is generally accepted to be a common event in human cancers, including OSCC.19 Rb is also a well-known tumor-suppressor, which functions through transition of the restriction point in progression of the cell cycle.
‘Endocytosis’ is responsible for bringing ligands, nutrients, plasma membrane proteins, and lipids into the cell interior and removing them from the cell surface. Oncogenic mutants of growth factor receptors show impaired endocytosis, and this peculiar endocytic system in cancer could be exploited by pharmacological therapies, to deliver effective patient treatment.20 Although endocytosis was characterized as an important pathway in OSCC, more studies are needed to clarify the mechanisms of this pathway in OSCC.
‘Pathways in cancer’ involves several signaling pathways and tumor-related genes, including Wnt, Ras, p53, and cell apoptosis. Aberrant activation of the Wnt pathway is highly associated with tumorigenesis and tumor metastasis.21 Ras is a small G protein that cycles between inactive GDP-bound and active GTP-bound forms, and functions as a molecular switch for signal transduction initiated in the cell membrane. Mutationally activated Ras proteins have been shown to induce nearly 30% of all human cancers.22 Cell apoptosis plays an important role in cancer, and de-regulation of apoptotic signaling and activation of anti-apoptotic systems allow cancer cells to evade apoptosis, leading to uncontrolled cell proliferation, tumor progression, therapeutic resistance, and cancer recurrence.23
Although the functions of some of the nine significant identified pathways have been investigated in OSCC, most have not. Further studies are therefore needed to clarify the roles of these other pathways, to provide new insights into the development and progression of OSCC.
This study had some limitations. Some of the lncRNAs identified by this method as being associated with OSCC remain to be validated. Furthermore, this method regarded all lncRNAs as equally important, and the extent of the association between individual lncRNAs and cancer were not considered.
In conclusion, we identified nine significant pathways that were regulated by a set of competitive lncRNAs in OSCC, suggesting that these lncRNAs may play important roles in the development of OSCC via the cell cycle and endocytosis.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Manikandan M, Deva Magendhra Rao AK, Arunkumar G, et al. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer 2016; 15: 28. DOI: 10.1186/s12943-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosebush MS, Rao SK, Samant S, et al. Oral cancer: enduring characteristics and emerging trends. J Mich Dent Assoc 2012; 94: 64–68. [PubMed] [Google Scholar]
- 3.Johnson NW, Warnakulasuriya S, Gupta PC, et al. Global oral health inequalities in incidence and outcomes for oral cancer: causes and solutions. Adv Dent Res 2011; 23: 237–246. DOI: 10.1177/0022034511402082. [DOI] [PubMed] [Google Scholar]
- 4.Brocklehurst PR, Baker SR, Speight PM. Oral cancer screening: what have we learnt and what is there still to achieve? Future Oncol 2010; 6: 299–304. DOI: 10.2217/fon.09.163. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136: 629–641. DOI: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015; 21: 1253–1261. DOI: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Yang YT, Wang YF, Lai JY, et al. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/beta-catenin signaling pathway. Cancer Sci 2016; 107: 1581–1589. DOI: 10.1111/cas.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Liu S, Cai G, et al. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci Rep 2015; 5: 15972. DOI: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J, Liu S, Sun Z, et al. LncRNAs2Pathways: identifying the pathways influenced by a set of lncRNAs of interest based on a global network propagation method. Sci Rep 2017; 7: 46566. DOI: 10.1038/srep46566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheu JJ, Lee CC, Hua CH, et al. LRIG1 modulates aggressiveness of head and neck cancers by regulating EGFR-MAPK-SPHK1 signaling and extracellular matrix remodeling. Oncogene 2014; 33: 1375–1384. DOI: 10.1038/onc.2013.98. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Fan X, Cai Y. A comparative study of improvements Pre-filter methods bring on feature selection using microarray data. Health Inf Sci Syst 2014; 2: 7. DOI: 10.1186/2047-2501-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumazin P, Yang X, Chiu HS, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 2011; 147: 370–381. DOI: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Kang S, Liu CC, et al. High-resolution functional annotation of human transcriptome: predicting isoform functions by a novel multiple instance-based label propagation method. Nucleic Acids Res 2014; 42: e39. DOI: 10.1093/nar/gkt1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Can T, Çamoǧlu O, Singh AK. Analysis of protein-protein interaction networks using random walks. Proceedings of the 5th International Workshop on Bioinformatics 2005; AMC: 1296–1300. DOI: 10.1080/00016480510012381.
- 15.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001; 125: 279–284. [DOI] [PubMed] [Google Scholar]
- 16.Hunt T. Nobel Lecture. Protein synthesis, proteolysis, and cell cycle transitions. Biosci Rep 2002; 22: 465–486. [DOI] [PubMed] [Google Scholar]
- 17.Ryan KM. p53 and autophagy in cancer: guardian of the genome meets guardian of the proteome. Eur J Cancer 2011; 47: 44–50. DOI: 10.1016/j.ejca.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758. DOI: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagpal JK, Patnaik S, Das BR. Prevalence of high-risk human papilloma virus types and its association with P53 codon 72 polymorphism in tobacco addicted oral squamous cell carcinoma (OSCC) patients of Eastern India. Int J Cancer 2002; 97: 649–653. [DOI] [PubMed] [Google Scholar]
- 20.Mellman I, Yarden Y. Endocytosis and cancer. Cold Spring Harb Perspect Biol 2013; 5: a016949. DOI: 10.1101/cshperspect.a016949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey N, Barwick BG, Moreno CS, et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer 2013; 13: 537. DOI: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol 2016; 58: 60–69. DOI: 10.1016/j.semcdb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammad RM, Muqbil I, Lowe L, et al. Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol 2015; 35: S78–S103. DOI: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]