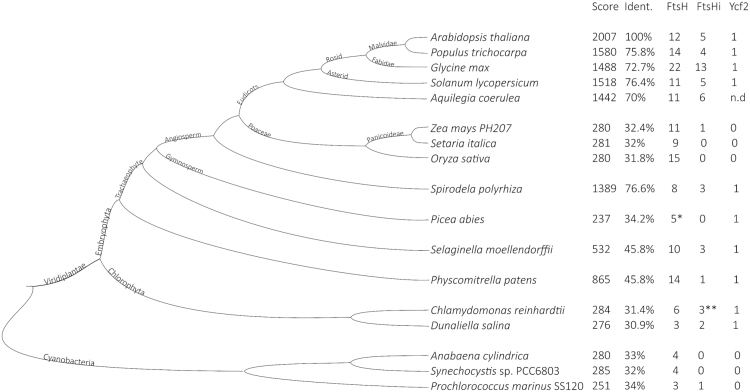
Fig. 1.
Co-evolution of FtsH12 and FtsHi. The FtsH12 amino acid sequence was blasted via Phytozome. Hits with the highest score and identity are shown. The number of presumably proteolytically active (FtsH) and -inactive (FtsHi) genes were determined by manually investigating the presence of an AAA-like domain and peptidase M41 domain. Sequences containing a zinc-binding motif (HEXXH) within M41 were assumed to correspond to proteolytically active proteases. Data for Picea abies were additionally retrieved from Congenie (marked with asterisks). Data for Prochlorococcus, Synechocystis, and Anabaena were retrieved from NCBI. Data for Chlamydomonas reinhardtii and Oryza sativa were supplemented with data from the literature (Garcia-Lorenzo et al., 2008, Malnoë et al., 2014). Ycf2 is plastid-encoded and therefore the search was performed on plastid-containing organisms, except for Aquilegia, in the NCBI organelle genome resources.