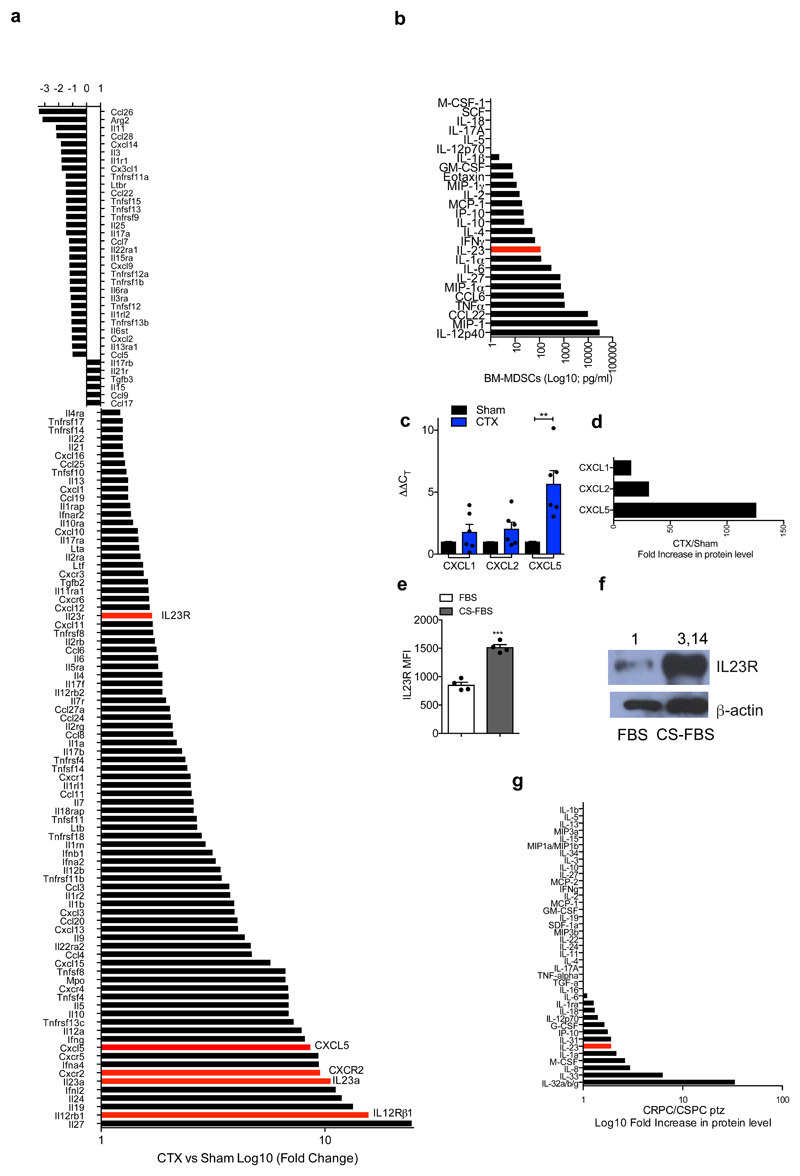
Extended Data 5. IL23 pathway is the most up-regulated in the tumor upon castration.
a, Gene expression of selected genes determined by NanoString nCounter gene expression assay in Sham Ptenpc-/- and CTX Ptenpc-/- tumors. Data are shown as pool of n=5. b, Analyses of the conditioned media of BM-derived MDSCs tested for the indicated soluble molecules by Mouse CytokineMAP B version 1.0. The graph reports the concentration of the indicated soluble molecules as Log10 of the concentration found in the conditioned medium of BM-MDSCs, the values were subtracted of the background (culture medium). Data are shown as pool of n=10. c, qRT-PCR analyses of the indicated genes in Sham (n=6) and CTX (n=6) Ptenpc-/- tumors. Data were reported as mean ± SEM of biological independent animals. Statistical analyses (Unpaired two sided t test): *P <0.05. d, Protein level of CXCL1, CXCL2 and CXCL5 in CTX Ptenpc-/- tumors. Data are analyzed as ratio between CTX (pool of 3 samples) and Sham (pool of 3 samples) Ptenpc-/- tumors and reported as fold increase in protein level. e-f, IL23R protein level analyzed by flow cytometry and western blot on TRAMP-C1 cells under normal culture condition (FBS) or androgen deprivation culture condition (CS-FBS). n=4 biological independent samples per group. f, Numbers indicate fold change in protein level. Loading control: anti–β–actin antibody. The WB was validated at twice. g, Protein profile of the plasma of CSPC and CRPC patients. Data are analyzed as ratio between CRPC (pool of 18 samples) and CSPC (pool of 17 samples) and reported as fold increase in protein level.