Abstract
MitoTracker ® dyes are fluorescent compounds that allow cellular mitochondrial content to be measured semi‐quantitatively by flow cytometry and have been used extensively in immunology publications. However, the parameters commonly reported, mean or median fluorescence intensity and percentage of cells that are MitoTracker® “high”, can be influenced by variability in cytometer setup, dye stability, and operator subjectivity, making it difficult to compare data between experiments. Here, we describe a method to identify MitoTracker® “high” populations in an objective manner. When analyzing data, we first removed outliers using a pre‐specified threshold, determined the fluorescence intensity of the brightest and dimmest events to obtain the fluorescence range and then gated cells within the top 90% of this range. This strategy substantially reduced variability between technical replicates and produced consistent results when data were analyzed by different operators. Consistent with previous reports and other analysis strategies, this analysis method demonstrated that within an individual, CD4+ T cells exhibit significantly higher mitochondrial mass than CD8+ T cells. Objective gating increases the reliability and utility of data generated using MitoTracker® dyes. © 2018 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: mitochondria, T‐lymphocytes, immunologic techniques, metabolism, flow cytometry
MitoTracker® probes are cell‐permeable, mitochondrion‐selective fluorescent dyes that accumulate in mitochondrial membranes 1. Two of the more popular probes are MitoTracker® Deep Red and MitoTracker® Green. MitoTracker® Deep Red binds thiol‐reactive chloromethyl groups in the mitochondrial membrane and is retained after cell fixation, whereas MitoTracker® Green binds free thiol groups of cysteine residues belonging to mitochondrial proteins and can only be used with non‐fixed cells. 2. In addition to fluorescence microscopy applications, MitoTracker® and other mitochondrial dyes are increasingly used in flow cytometry to provide a semi‐quantitative measure of mitochondrial mass and activity at the cellular level. Higher intensity staining of T cells with MitoTracker® dyes is associated with greater cellular mitochondrial content and increased respiration (as measured by oxygen consumption rate (OCR) and spare respiratory capacity (SRC)) 3, 4, 5, 6. Flow cytometry is a high‐throughput method of studying T cells, and MitoTracker® dyes can be combined with fluorescent antibodies to yield a detailed phenotypic profile of cell populations of interest. This approach has been used to study T cell mitochondria in animal models, and in human studies of diseases such as cancer, HIV, and HBV infection 7, 8, 9, 10, 11. However, for data to be comparable between independent experiments, care must be taken to ensure the consistency of the fluorescent signal. This is particularly relevant for clinical studies because testing, which is typically performed on freshly isolated cells, may take place over weeks, months, or even years. We submit that the conventional methods used to assess the MitoTracker® fluorescence signal are unsuitable in this context.
The most commonly reported metric of MitoTracker® fluorescence is mean or median fluorescence intensity (MFI) 7, 9. This approach is unbiased and is appropriate for comparing between cell populations within an individual test. However, for the MFI of any fluorescence parameter to be comparable between experiments, consistent cytometer settings and/or calibration beads must be used; few if any of the studies published to date report including these measures in their experimental protocols. Even if consistent settings are used, we demonstrate here that different aliquots of dye from the same lot can vary in fluorescence intensity, and may lose stability over time even if stored as directed. For these reasons, MFI cannot reliably be used to compare MitoTracker® staining between experiments conducted at different times.
The second reported method is to gate MitoTracker® “high” populations by human eye, but this method is unavoidably subjective 12. All T cells contain mitochondria and so conventional cytometry controls such as fluorescence minus one (FMO) controls are not helpful in this context. This method will therefore produce varying results depending on the operator performing the analysis.
Here, we present a novel method to allow the objective delineation of MitoTracker® “high” populations.
Materials and Methods
Ethics Statement
All participants provided written informed consent. Experimental protocols were approved by the University of North Carolina Institutional Biomedical Review Board (IRB number 11–1506) and carried out in accordance with the relevant guidelines. All procedures were in accordance with the Helsinki Declaration of 1975, as revised in 2008.
Antibodies and Fluorescent Dyes
Zombie NIR Fixable Live/Dead cell stain (BioLegend) was used to assess cell viability. The dye was reconstituted using DMSO (Sigma‐Aldrich) and stored at −20°C. The following cell surface antibodies were used: CD3‐PE‐Cy7, CD4‐Brilliant Violet 421, CD8‐Brilliant Violet 510, CD14‐Brilliant Violet 650, CD16‐Brilliant Violet 650, CD19‐Brilliant Violet 650, and CD56‐Brilliant Violet 650 (all BioLegend). Antibodies were stored at 4°C. Stock solutions of 1 mM MitoTracker® Deep Red (MT Deep Red or MTDR; Molecular Probes lot 1,837,994) and 1 mM MitoTracker® Green (MT Green or MTG; Molecular Probes lot 1,842,298) were prepared by adding DMSO to vials of lyophilized dye. Single‐use aliquots were stored at −20°C. All refrigerators and freezers were electronically temperature‐monitored and did not fall outside their acceptable temperature ranges during the period of these experiments.
Cell Isolation and Staining
Peripheral blood mononuclear cells (PBMC) were isolated from whole blood by Ficoll–Hypaque density gradient centrifugation using SepMate tubes (STEMCELL Technologies). Freshly‐isolated PBMC (5 × 105 per condition) were stained in a volume of 100 μl PBS with Zombie NIR Fixable Live/Dead cell stain for 20 min at room temperature, followed by surface antibodies in a total volume of 100 μl PBS/2% FBS for 30 min at room temperature. Finally, cells were incubated in 200 μl warm PBS containing 15 nM MT Deep Red and 50 nM MT Green (unless otherwise indicated) for 1 h at 37°C. This extended staining period improved data reproducibility relative to staining for 30 min (data not shown). To ensure no spectral overlap into the MT Deep Red and MT Green channels, fluorescence minus one (FMO) controls containing all fluorochromes except the dye of interest were used. Compensation controls were prepared for each fluorochrome using anti‐mouse Ig κ compensation particles (BD Biosciences), with the exception of Zombie, MT Deep Red and MT Green, where single‐stained cells were used for compensation. Samples were acquired, unfixed unless otherwise specified, by flow cytometry within 2 h of the completion of staining. Where indicated, cells were fixed in PBS/2% paraformaldehyde for 30 min prior to acquisition. A minimum of 20,000 T cells were acquired for analysis.
Flow Cytometry
Samples were acquired using an LSRFortessa flow cytometer equipped with FACSDiva software version 8.0.1 (BD Biosciences). Cytometer calibration was performed daily using CS&T Research Beads (BD Biosciences). Application settings were created in FACSDiva and used in each experiment to ensure consistent fluorochrome excitation.
Data were analyzed using FlowJo version 10.4.2 (Tree Star). Fluorescence parameters were compensated in FlowJo using the compensation controls. CD4+ and CD8+ T cells were identified using a hierarchical gating strategy.
Imaging Flow Cytometry
PBMC were stained with CD3‐PerCP‐Cy5.5 (Biolegend), 50 nM MitoTracker® Green and 15 nM MitoTracker® Deep Red and acquired on an Amnis ImageStreamX Mark II. Data were analyzed using IDEAS version 6.2 (Amnis).
Analysis Method
Hypothesis
Our method for gating MitoTracker® “high” populations is based on the observation that despite variations in MFI and fluorescence range between different dye aliquots, the distribution of events within the fluorescence range appeared similar. This suggested that the percentage of cells lying within a defined fraction of this range (e.g., the top 90% of the range) would remain constant.
A mathematical justification for our hypothesis that the probability of a cell lying within a specified subrange (e.g., the top 90% of the range) is the same for different dye aliquots is detailed as follows:
Let X denote a random variable corresponding to the fluorescence value when using a particular dye aliquot. Assume the distribution of X has finite support, with lower and upper bounds l x and u x, respectively. The probability of a cell lying within the top 90% of the support is then Pr[u x − 0.9 r x < X < u x] where r x = u x − l x is the range of the finite support, and Pr[.] denotes probability.
Assume using a different dye aliquot results in a linear transformation of the fluorescence value (see Fig. 2C for data supporting this assumption). Specifically, if a different dye aliquot is used, let Y denote a random variable corresponding to the fluorescence value of that dye aliquot and Y = a X + b, for constants a and b. It follows that the distribution of Y has finite support, with lower and upper bounds l y and u y, respectively, where l y = a l x + b and u y = a u x + b. Moreover, using this dye aliquot, the probability of a cell lying within the top 90% of the support is Pr[u y − 0.9 r y < Y < u y] where r y = u y – l y. Note r y = a (u x – l x) = a r x so that Pr[u y − 0.9 r y < Y < u y] = Pr[a u x + b – 0.9 a r x < a X + b < a u x + b] = Pr[a (u x – 0.9 r x) + b < a X + b < a u x + b]. This simplifies to Pr[u x – 0.9 r x < X < u x], that is, the probability of a cell lying within the top 90% of the support is the same for different dye aliquots.
Figure 2.
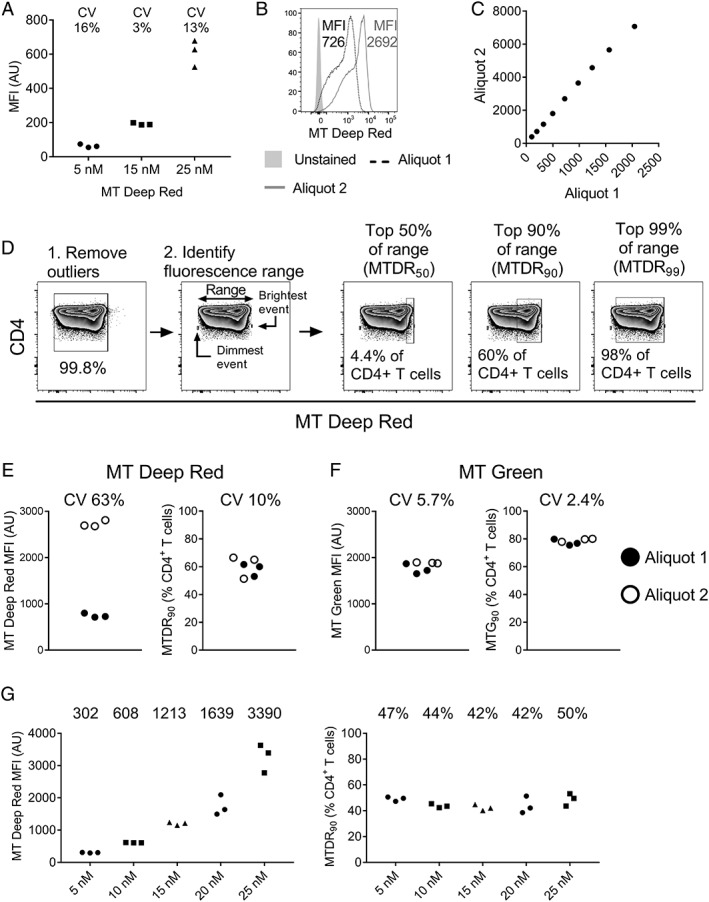
(A) Median fluorescence intensity (MFI) of MitoTracker® deep red in CD4+ T cells exposed to different concentrations of the dye (three technical replicates per concentration). Data are representative of two independent experiments in different donor PBMC. (B) MT deep red fluorescence of CD4+ T cells from the same donor stained simultaneously with two different aliquots of dye from the same lot (15 nM concentration). (C) Quantile‐quantile plot demonstrating a linear relationship between the population distributions of cells stained with the two dye aliquots. The fluorescence intensity of the 10th, 20th, …, 90th percentiles are plotted. (D) Strategy for the objective gating of MT deep red CD4+ T cell populations. After removing outliers, the fluorescence of the dimmest and brightest events was determined and the dimmest fluorescence was subtracted from the brightest fluorescence to obtain the trimmed fluorescence range. The percentage of cells in the top 50% of the range (MTDR50), the top 90% of the range (MTDR90), and the top 99% of the range (MTDR99) were gated as shown. For detailed experimental protocol, see Methods. (E, F) Inter‐aliquot variability of MT deep red (E) and MT green (F) using MFI (left) compared with objective gating (right). (G) The effect of using different doses of MT deep red on CD4+ T cell MFI (left) and the percentage of cells within the top 90% of the fluorescence range (MTDR90; right). Numbers represent the median value at each dye concentration. AU, arbitrary units; CV, coefficient of variation; MFI, median fluorescence intensity; MT, MitoTracker®.
In practice, the lower and upper bounds of fluorescence intensity (l x and u x) are not known, but rather need to be estimated. In the absence of measurement error, the minimum and maximum observed intensities could be used to estimate l x and u x. However, suppose due to measurement error that the true fluorescence value for a particular dye is not observed, but rather we observe the true value plus some independent noise. That is, instead of observing X, suppose we observe X + e where e is a random variable representing measurement error which has mean zero and is independent of X. In this case, the minimum and maximum observed intensities would be expected to yield biased estimates of l x and u x. Assuming the variance of e is small relative to the range of the support of X, then removing outliers to obtain a trimmed range (see below) would be expected to yield more accurate estimates of l x and u x.
Gating method
In accordance with this hypothesis, MT Deep Red “high” CD4+ or CD8+ T cells were gated as follows: first, to remove outliers that might skew the positioning of the gate, the top 0.1% and the bottom 0.1% of cells in terms of MT Deep Red brightness were excluded, leaving 99.8% of the total (the “trimmed” cell population). Next, the brightest remaining cell and the dimmest remaining cell were individually gated (or one of the brightest and dimmest cells if there were multiple cells with exactly the same fluorescence intensity) and the fluorescence intensity was obtained for each. The dimmest fluorescence intensity was subtracted from the brightest to obtain the trimmed fluorescence range. Then, 90% of this range was identified by multiplying the fluorescence range by 0.9:
A gate was placed manually between this value and the brightest event. The percentage of cells falling within this gate was designated the “MTDR90” fraction (Fig. 2D). In selected experiments, the percentage of cells lying in the top 99% (MTDR99) and the top 50% (MTDR50) of the fluorescence range were also determined. The same methodology was used to identify MT Green “high” populations. Independent gates were defined for CD4+ and CD8+ T cell populations as the fluorescence range differs between these subsets (see Fig. 3).
Figure 3.
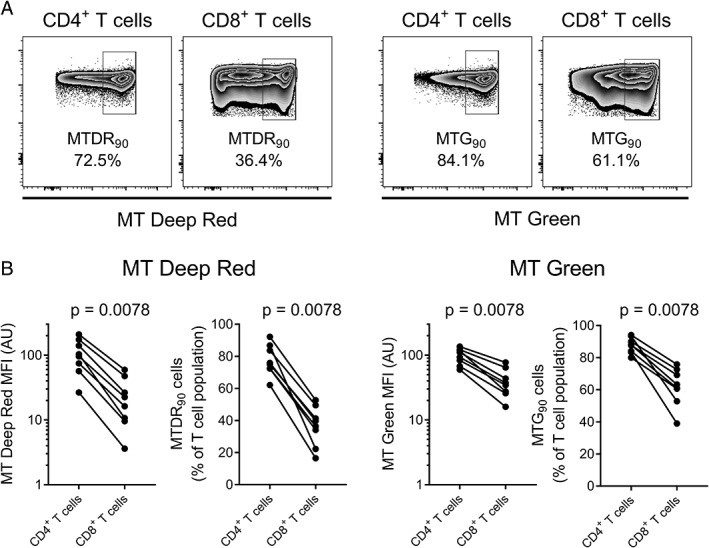
MitoTracker® staining is brighter in CD4+ T cells than CD8+ T cells. (A) MT deep red and MT green staining was compared between CD4+ and CD8+ T cells from the same individual (n = 8) using both MFI and the objective gating method (cells in the top 90% of the fluorescence range, or MT90). (B) By both methods, MitoTracker® fluorescence was consistently higher in CD4+ T cells than CD8+ T cells. Data analyzed using Wilcoxon matched‐pairs signed rank test. AU, arbitrary units; MFI, median fluorescence intensity; MT, MitoTracker®.
Statistical Analyses
Data were analyzed using GraphPad Prism Version 7. Outcomes were compared between paired samples using an exact Wilcoxon matched‐pairs signed rank test. Correlations were performed using Spearman's rank correlation. Statistical testing was two‐sided and P‐values < 0.05 were considered statistically significant.
Results
Variability of MT Deep Red Fluorescence Intensity between Batches from the Same Lot
MitoTracker® dyes have previously been reported to bind specifically to mitochondria in respiring cells 2. Using imaging flow cytometry, we confirmed that cells that are MitoTracker® “high” by conventional flow cytometry (gated by human eye) displayed increased intracellular staining with MT Deep Red and MT Green relative to MitoTracker® “low” cells, with clear colocalization between the two dyes (Supporting Information Fig. 1).
We next determined the optimal concentration of MT Deep Red to achieve reproducibility between technical replicates. PBMC were stained in triplicate with either 5 nM, 15 nM, or 25 nM MT Deep Red. The gating strategy used to identify CD4+ and CD8+ T cells is shown in Figure 1. As expected, median fluorescence intensity (MFI) increased with higher dye concentration (Fig. 2A and Supporting Information Fig. 2). The inter‐replicate variation (as measured by coefficient of variation, CV) was lowest with 15 nM MT Deep Red; accordingly, this concentration was used for subsequent experiments.
Figure 1.
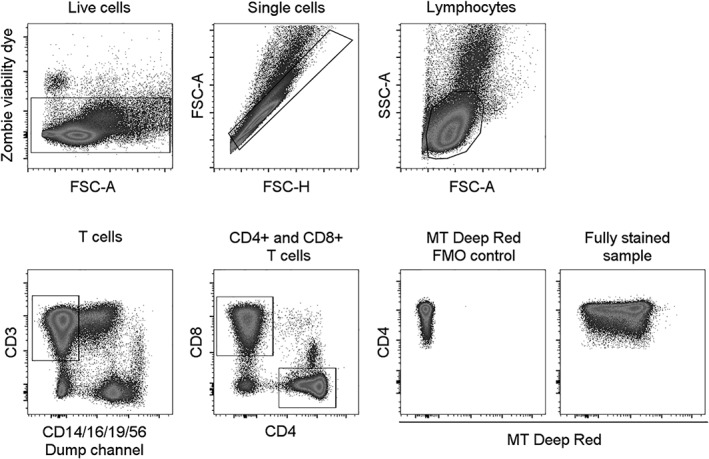
Gating strategy to identify CD4+ and CD8+ T cells. FMO, fluorescence minus one; FSC‐A, forward scatter area; FSC‐H, forward scatter height; SSC‐A, side scatter area; MT, MitoTracker®.
To ensure reproducibility over time, it is necessary to be confident that different aliquots of dye produce consistent results. To assess the consistency of MT Deep Red MFI, PBMC from the same donor were stained simultaneously in triplicate with dye from two single‐use aliquots of the same lot. We observed considerable variation in MFI following staining with the two aliquots, with one aliquot giving an MFI almost four times higher than the other for CD4+ T cells despite both having been prepared together and stored as directed by the manufacturer (Fig. 2B,E). We concluded that it would not be possible to use MFI to compare MitoTracker® staining between experiments.
Gating MitoTracker® Populations Using Fluorescence Range Substantially Improves Reproducibility
We observed that while there was substantial variation between aliquots in MitoTracker® fluorescence in terms of MFI (and the minimal and maximal fluorescence intensity), the distribution of events within the fluorescence range appeared similar (Fig. 2B). To assess whether different aliquots result in a linear transformation of the distribution of fluorescence intensity of cells, we performed a quantile‐quantile analysis, comparing the fluorescence intensity of cells at the 10th, 20th, 30th … 90th percentiles, and observed a clear linear relationship between aliquots (Fig. 2C). This led us to hypothesize that the percentage of cells lying within a defined percentage of this fluorescence range would remain constant, and that a strategy that objectively gated on this percentage could provide more reproducible results than MFI‐based gating. Using this approach (see details in Methods section), MitoTracker® Deep Red outliers were removed, the fluorescence range of the remaining cells was determined, and cells lying within the top 50%, the top 90%, and the top 99% of this fluorescence range were gated (Fig. 2D). The population falling within the top 90% of the fluorescence range, designated MTDR90, best reflected the “mitochondrial dye high” population described in previous publications 9, 12, 13. The variability between technical replicates using MTDR90 gating was substantially lower than the variability in MT Deep Red MFI (10% vs. 63% CV). (Fig. 2E).
To determine whether objective gating also improved consistency with other mitochondrial dyes, we compared staining with two aliquots of MitoTracker® Green using MFI and gating the top 90% of the fluorescence range (MTG90). While the variability between aliquots is typically lower for MT Green than for MT Deep Red, we show that fluorescence range gating further improved assay reproducibility (MTG90 CV 2.4% compared with 5.7% when using MFI) (Fig. 2F). For both MitoTracker® Deep Red and MitoTracker® Green, highly reproducible results were obtained when the data were analyzed by three independent operators (Supporting Information Fig. 3).
Next, we assessed whether MT90 gating produced consistent results over a range of MitoTracker® concentrations by staining PBMC with MT Deep Red doses ranging from 5 nM to 25 nM. As expected, MT Deep Red MFI increased in CD4+ T cells with increasing dye concentration (Fig. 2G, left graph). However, the percentage of CD4+ T cells in the top 90% of the fluorescence range (MTDR90) was similar across dye concentrations (Fig. 2G, right graph), confirming that the distribution of cells within the “high” fluorescence subrange remains constant when dye brightness increases. Fluorescence range gating can thus be used to compare results from experiments where dye fluorescence is variable. In a separate experiment, we confirmed that MTDR90 gating also produces consistent results with different dye concentrations when cells are fixed prior to acquisition (Supporting Information Fig. 4).
Objective Gating Recapitulates Comparisons between CD4+ and CD8+ T Cells Made Using MFI
To further validate this method, we compared MT Deep Red and MT Green staining between CD4+ and CD8+ T cells from eight blood donors, using both objective gating and MFI. As the comparison is between cells within an individual stained simultaneously, MFI is a valid measure in this instance. As shown in Figure 3, the MT Deep Red and MT Green signal was higher in CD4+ than CD8+ T cells in all eight individuals, both in terms of MFI and the percentage falling within the top 90% of the fluorescence range (MTDR90 or MTG90), reproducing previous observations 6, 8. We also observed significant differences between CD4+ and CD8+ T cells when cells falling within the top 50% of the fluorescence range (MT50) and the top 99% of the fluorescence range (MT99) were gated (Supporting Information Fig. 5). However, MT90 gating produced the strongest correlation with MFI in terms of the fold‐change increase in CD4+ T cells relative to CD8+ T cells (Table 1). This suggested that MT90 gating best discriminates between different T cells subsets and is consistent with our observation that MT90 gating best identified the previously described MitoTracker® “high” population.
Table 1.
Both MT90 and MT50 gating show significant correlation with MFI when comparing CD4+ and CD8+ T cells in the same participant
| MitoTracker® Deep Red | MitoTracker® Green | |||||
|---|---|---|---|---|---|---|
| MTDR50 vs. MFI | MTDR90 vs. MFI | MTDR99 vs. MFI | MTG50 vs. MFI | MTG90 vs. MFI | MTG99 vs. MFI | |
| Spearman r | 0.79 | 0.83 | 0.69 | 0.83 | 0.88 | 0.59 |
| P value | 0.028 | 0.015 | 0.069 | 0.015 | 0.007 | 0.134 |
To examine the correlation between MT gating and MFI, the within‐participant fold‐change in MitoTracker® MFI in CD4+ T cells compared with CD8+ T cells (CD4 MFI/CD8 MFI) was calculated and then correlated with the fold‐change (CD4 MT/CD8 MT) in MT50, MT90, and MT99 for each sample shown in Figure 3. Correlations performed using Spearman's rank correlation. Bold text indicates significant correlations (two‐tailed P value <0.05).
Discussion
We have described a novel, objective method to identify MitoTracker® “high” T cell populations, circumventing the problems presented by existing analytical approaches. As we have shown, objective gating delivers consistent results even in the face of differences in dye fluorescence intensity, overcoming the major limitation of comparing MFI between experiments. Second, in contrast to gating by human eye, our method delivers an unbiased value for the percentage of MitoTracker® “high” cells. This approach affords consistent results that are operator‐independent.
Even using this approach, we recommend using consistent cytometer settings and/or calibration beads to ensure consistent cytometer performance between experiments and to eliminate operator‐introduced variability when performing the cytometer setup. It is also advisable to perform experiments in triplicate (or more) to assess the consistency of staining, and so that any anomalous results can be identified and excluded. We note that experiments involving compounds such as FCCP that reduce mitochondrial polarization in all cells will not be amenable to analysis using this method.
Although we have focused on MitoTracker® dyes in this report, objective gating could be used to gate populations stained with other mitochondrial dyes, and with other fluorescent antibodies or cellular dyes where delineating “high” and “low” populations is challenging. For example, multiple studies have reported differences in function and response to immunotherapy between PD‐1 “high” and PD‐1 “intermediate” or “low” T cells, but as yet there is no standardized method for identifying these different populations 14, 15, 16. As we have demonstrated, the method can be modified to gate, for example, cells in the top 50% of the fluorescence range, according to the requirements of the researcher.
Ex vivo metabolic measurements are typically performed using freshly‐isolated cells 4, 12, 17. Therefore, longitudinal assessment of metabolic activity such as mitochondrial mass and function is challenging because of intra‐lot variability of cellular dyes. Here, we propose an approach that enables more reproducible measurement of ex vivo mitochondrial mass and activity that is operator independent and overcomes limitations associated with intra‐lot variability of dyes. Robust, reproducible data using mitochondrial dyes could provide valuable insights into the role of mitochondria in human health and disease.
Supporting information
Appendix S1: Supporting information
Supplementary Figure 1: A) Gating strategy for the identification MitoTracker® “‘high”’ and “‘low”’ subsets of focused single CD3+ T cells. B) Compensated images of MitoTracker® Green “‘high”’ and “‘low”’ cells obtained by imaging flow cytometry at 60x magnification. C) Single images of MT Green ‘high’ and ‘low’ and MT Deep Red ‘high’ and ‘low’ T cells.
Supplementary Figure 2: Flow cytometry plots of CD4+ T cells stained in triplicate with three different concentrations of MitoTracker® Deep Red. MT, MitoTracker®.
Supplementary Figure 3: Analysis by three independent operators of the percentage of CD4+ T cells in the top 90% of the fluorescence range for MitoTracker® Deep Red (MTDR90; left) and MitoTracker® Green (MTG90; right). Data from operator 1 are reproduced from Figure 2E and F.
Supplementary Figure 4: The effect of using different doses of MitoTracker® Deep Red on CD4+ T cell MFI (left) and the percentage of cells within the top 90% of the fluorescence range (MTDR90; right) when cells are fixed with 2% paraformaldehyde after staining. Numbers represent the median value at each dye concentration. AU, arbitrary units; MFI, median fluorescence intensity; MT, MitoTracker®.
Supplementary Figure 5: Comparison of MT Deep Red and MT Green staining between CD4+ and CD8+ T cells from the same individual (n = 8) gating cells in the top 99% of the fluorescence range (MT99) or cells in the top 50% of the range (MT50). Data analyzed using Wilcoxon matched‐pairs signed rank test. MT, MitoTracker®.
Acknowledgments
We thank Dr. Richard Dunham for reviewing the manuscript and providing helpful comments, Dr. Maria Abad and Yinyan Xu for performing independent analysis of these data, and Sébastien Coquery for assistance with imaging flow cytometry. This study was supported by NIH NIAID grant U01 AI131310. Research reported in this publication was supported by the UNC Center for AIDS Research award number P30AI050410 and by the Office of the Director, National Institutes of Health under award number 1 S10 OD017984‐01A1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The UNC Flow Cytometry Core Facility is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Literature Cited
- 1. Metivier D, Dallaporta B, Zamzami N, Larochette N, Susin SA, Marzo I, Kroemer G. Cytofluorometric detection of mitochondrial alterations in early CD95/Fas/APO‐1‐triggered apoptosis of Jurkat T lymphoma cells. Comparison of seven mitochondrion‐specific fluorochromes. Immunol Lett 1998;61:157–163. [DOI] [PubMed] [Google Scholar]
- 2. Cottet‐Rousselle C, Ronot X, Leverve X, Mayol JF. Cytometric assessment of mitochondria using fluorescent probes. Cytometry Part A 2011;79A:405–425. [DOI] [PubMed] [Google Scholar]
- 3. Fischer M, Bantug GR, Dimeloe S, Gubser PM, Burgener AV, Grählert J, Balmer ML, Develioglu L, Steiner R, Unterstab G, et al. Early effector maturation of naive human CD8(+) T cells requires mitochondrial biogenesis. Eur J Immunol 2018;48:1632–1643. [DOI] [PubMed] [Google Scholar]
- 4. Henson SM, Lanna A, Riddell NE, Franzese O, Macaulay R, Griffiths SJ, Puleston DJ, Watson AS, Simon AK, Tooze SA, et al. p38 signaling inhibits mTORC1‐independent autophagy in senescent human CD8(+) T cells. J Clin Invest 2014;124:4004–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 2016;166:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao Y, Rathmell JC, Macintyre AN. Metabolic reprogramming towards aerobic glycolysis correlates with greater proliferative ability and resistance to metabolic inhibition in CD8 versus CD4 T cells. PLoS One 2014;9:e104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schurich A, Pallett LJ, Jajbhay D, Wijngaarden J, Otano I, Gill US, Hansi N, Kennedy PT, Nastouli E, Gilson R, et al. Distinct metabolic requirements of exhausted and functional virus‐specific CD8 T cells in the same host. Cell Rep 2016;16:1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu F, Hao Y, Zhao H, Xiao J, Han N, Zhang Y, Dai G, Chong X, Zeng H, Zhang F. Distinct mitochondrial disturbance in CD4+T and CD8+T cells from HIV‐infected patients. J Acquir Immune Defic Syndr 2017;74:206–212. [DOI] [PubMed] [Google Scholar]
- 9. Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, Chiang YCJ, Corona AL, Gemta LF, Vincent BG, et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2017;2 https://www.ncbi.nlm.nih.gov/pubmed/28614802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM, et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD‐1 are an early driver of CD8+ T cell exhaustion. Immunity 2016;45:358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fisicaro P, Barili V, Montanini B, Acerbi G, Ferracin M, Guerrieri F, Salerno D, Boni C, Massari M, Cavallo MC, et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV‐specific CD8 T cells in chronic hepatitis B. Nat Med 2017;23:327–336. [DOI] [PubMed] [Google Scholar]
- 12. Petrovas C, Mueller YM, Dimitriou ID, Altork SR, Banerjee A, Sklar P, Mounzer KC, Altman JD, Katsikis PD. Increased mitochondrial mass characterizes the survival defect of HIV‐specific CD8(+) T cells. Blood 2007;109:2505–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bettonville M, D'Aria S, Weatherly K, Porporato PE, Zhang J, Bousbata S, Sonveaux P, Braun MY. Long‐term antigen exposure irreversibly modifies metabolic requirements for T cell function. eLife 2018;7 https://www.ncbi.nlm.nih.gov/pubmed/28614802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 2012;338:1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blackburn SD, Shin H, Freeman GJ, Wherry EJ. Selective expansion of a subset of exhausted CD8 T cells by αPD‐L1 blockade. Proc Natl Acad Sci USA 2008;105:15016–15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C, et al. A transcriptionally and functionally distinct PD‐1(+) CD8(+) T cell pool with predictive potential in non‐small‐cell lung cancer treated with PD‐1 blockade. Nat Med 2018;24:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keane KN, Calton EK, Cruzat VF, Soares MJ, Newsholme P. The impact of cryopreservation on human peripheral blood leucocyte bioenergetics. Clin Sci (Lond) 2015;128:723–733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information
Supplementary Figure 1: A) Gating strategy for the identification MitoTracker® “‘high”’ and “‘low”’ subsets of focused single CD3+ T cells. B) Compensated images of MitoTracker® Green “‘high”’ and “‘low”’ cells obtained by imaging flow cytometry at 60x magnification. C) Single images of MT Green ‘high’ and ‘low’ and MT Deep Red ‘high’ and ‘low’ T cells.
Supplementary Figure 2: Flow cytometry plots of CD4+ T cells stained in triplicate with three different concentrations of MitoTracker® Deep Red. MT, MitoTracker®.
Supplementary Figure 3: Analysis by three independent operators of the percentage of CD4+ T cells in the top 90% of the fluorescence range for MitoTracker® Deep Red (MTDR90; left) and MitoTracker® Green (MTG90; right). Data from operator 1 are reproduced from Figure 2E and F.
Supplementary Figure 4: The effect of using different doses of MitoTracker® Deep Red on CD4+ T cell MFI (left) and the percentage of cells within the top 90% of the fluorescence range (MTDR90; right) when cells are fixed with 2% paraformaldehyde after staining. Numbers represent the median value at each dye concentration. AU, arbitrary units; MFI, median fluorescence intensity; MT, MitoTracker®.
Supplementary Figure 5: Comparison of MT Deep Red and MT Green staining between CD4+ and CD8+ T cells from the same individual (n = 8) gating cells in the top 99% of the fluorescence range (MT99) or cells in the top 50% of the range (MT50). Data analyzed using Wilcoxon matched‐pairs signed rank test. MT, MitoTracker®.


